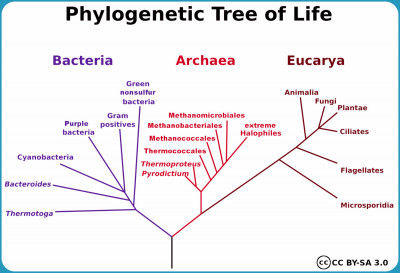
Terms
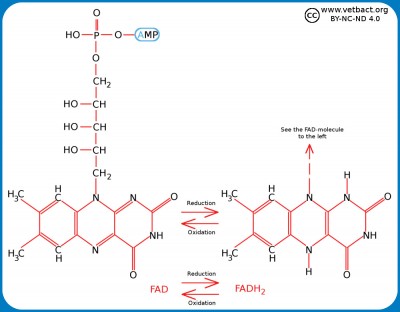
Adenosine triphosphate (ATP)
Structural formula for ATP. The three phosphate groups are named alpha, beta and gamma from the ribose moiety. The nitrogen base (adenine) together with the sugar moiety (ribose) forms the molecule adenosine. If adenosine has only one phosphate group, it is called adenosine monophosphate (AMP). - Click on the image to enlarge it.
Introduction
Adenosine triphosphate (ATP) is a nucleotide (nucleoside triphosphate) consisting of a nitrogen base (adenine), a sugar molecule (ribose) and a triphosphate group. ATP is the most important energy form for all organisms and can be said to be the cell's energy currency. The reason for this is that ATP contains energy-rich bonds between the phosphate groups and the chemical energy in these bonds can be recovered when such a group is cleaved from ATP and then ADP (adenosine diphosphate) is formed or from ADP to AMP (adenosine monophosphate). The cell cannot store ATP and the synthesis of ATP must constantly be regulated in relation to the need. ATP is also a building block for synthesis of nucleic acid.
Formation of ATP
Energy is required for the synthesis of ATP from ADP and phosphate. That energy can be generated by glycolysis, during the Krebs cycle and through the electron transport chain. Some intracellular bacteria (e.g. members of the Chlamydiaceae family) cannot synthesize the amount of ATP required for the various functions of the cell and must, therefore, parasitize on the host cell's ATP synthesis.
Use of energy in ATP
The energy-demanding processes, which utilize ATP, can in bacterial cells be divided into three main groups:
- Mechanical work (eg flagella rotation)
- Active transport (i.e., movement of molecules across membranes and towards a concentration gradient)
- Konstruction of molecules from their building blocks (i.e.synthesis of eg DNA and proteins).
Other
The German-British biochemist Hans Krebs was awarded the Nobel Prize in physiology or medicine in 1953 for investigating and presenting the most important parts of the citric acid cycle in the 1930s.
Updated: 2019-10-17.
Adhesion (Adherence)
Introduction
Adhesion in bacteriological contexts refers to the ability of bacteria to stick to surfaces (e.g. tissues or foreign material i.e. prostheses). This ability can be more or less specific. Colonization of bacteria to a surface and the formation of a biofilm begins with adhesion to the surface. If the surface consists of an inert foreign material, the adhesion is usually non-specific.
Specific adhesion
For specific adhesion to take place, there must be a receptor on the surface and a ligand on the bacterium, which can bind to the surface. Bacteria use the so-called adhesins as ligands to attach to different receptors on cells in the host animal tissue. Adhesins are proteins or polysaccharides and their receptors can also be made of proteins or polysaccharides. Adhesins are important virulence factors because they contribute to the ability of bacteria to colonize different tissues. Many pathogenic bacteria can express a wide variety of different adhesins, which are expressed during different phases of the infection. Inhibition of one of these adhesins usually results in the bacterium losing its pathogenicity. Attempts to use anti-adhesin antibodies as vaccines have in some cases been successful in model systems.
A well-characterized bacterial adhesin is FimH, which is bound to pili and produced by uropathogenic Escherichia coli (UPEC). FimH binds to mannose and CD48 is a type of human immunoglobulin, which is glycosylated and contains mannose residues. TLR4 is a member of the so-called the toll-like- receptor family, which consists of human membrane proteins that are important for innate immunity. The image is adapted from Wikipedia, Bacterial adhesion. - Click on the image to enlarge it.
Adhesin-receptor systems in bacteria
Fimbriae (= common pili) often function as adhesins and sometimes a particular polypeptide, which is localized at the tip of the fimbria is the actual ligand. Membrane proteins may also act as adhesins.
Unspecifik adhesion
Many bacteria produce extracellular polysaccharides, which constitute unspecific adhesion factors. The extracellular polysaccharide poly-N-acetylglucosamine (PNAG) works e.g. as an adhesion factor for some staphylococci and allows them to stick to certain abiotic (non-living) surfaces and form a biofilm there.
Reference
Di Martino, P. 2018. Bacterial adherence: Much more than a bond. AIMS Microbiol. 4(3): 563-566.
Updated: 2023-05-31.
Anaerobic bacteria
Introduction
Anaerobic bacteria are bacteria that cannnot use oxygen in their metabolism, but are poisoned and killed by this molecule. These bacteria are also said to be strictly anaerobic (= obligate anaerobic) to distinguish them from the oxygen tolerant bacteria and the facultatively anaerobic (= facultatively aerobic) bacteria. Oxygen tolerant bacteria does not use oxygen in their metabolism. However, they are not poisoned by oxygen, but can live and multiply in the presence of oxygen, at least for a certain time. Facultatively anaerobic bacteria are not poisoned by oxygen and can switch their metabolism, so that in the presence of oxygen they utilize oxygen in metabolism, but in the absence of oxygen, they can extract energy in other ways (e.g. by fermentation or anaerobic respiration).
How can oxygen be toxic for cells?
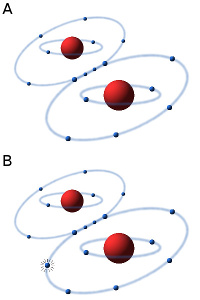 The oxygen molecule consists of two atoms of oxygen, making the molecule stable because the two atoms can then share a pair of electrons in the outer electron shell (see Fig. A). In the presence of oxygen (O2) in an aqueous solution (e.g. in a cell) small amounts of hydrogen peroxide (H2O2) and superoxide radicals are always formed by an equilibrium reaction. The superoxide radical (see Fig. B) is usually designated O2·-, to show that it consists of an oxygen molecule, which has taken up an extra electron (·), making it negatively charged. Hydrogen peroxide and the superoxide radical in particular are highly toxic to cells because they are very reactive and can affect a variety of substances, e.g. oxidize unsaturated fatty acids, leading to the so-called oxidative stress. In order to survive in an oxygen-containing environment, the cells, therefore, have enzymes that can metabolize (detoxify) hydrogen peroxide and the superoxide radical.
The oxygen molecule consists of two atoms of oxygen, making the molecule stable because the two atoms can then share a pair of electrons in the outer electron shell (see Fig. A). In the presence of oxygen (O2) in an aqueous solution (e.g. in a cell) small amounts of hydrogen peroxide (H2O2) and superoxide radicals are always formed by an equilibrium reaction. The superoxide radical (see Fig. B) is usually designated O2·-, to show that it consists of an oxygen molecule, which has taken up an extra electron (·), making it negatively charged. Hydrogen peroxide and the superoxide radical in particular are highly toxic to cells because they are very reactive and can affect a variety of substances, e.g. oxidize unsaturated fatty acids, leading to the so-called oxidative stress. In order to survive in an oxygen-containing environment, the cells, therefore, have enzymes that can metabolize (detoxify) hydrogen peroxide and the superoxide radical.
Detoxification of hydrogen peroxide and superoxide radicals
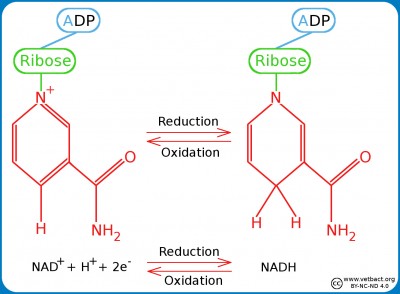
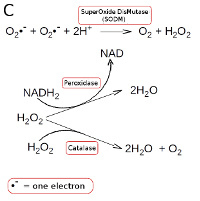 Strictly anaerobic bacteria lacks all enzymes which metabolize hydrogen peroxide and superoxide radicals. Other bacteria have superoxide dismutase (SODM), which converts superoxide radicals into hydrogen peroxide and oxygen (see Fig. C). Aerobic and most facultatively anaerobic bacteria have catalase, which converts hydrogen peroxide to water and oxygen (see Fig. C). Many oxygen-tolerant anaerobic bacteria have a peroxidase, which converts hydrogen peroxide to water by utilizing NADH2 (see Fig. C; click on it to enlarge it).
Strictly anaerobic bacteria lacks all enzymes which metabolize hydrogen peroxide and superoxide radicals. Other bacteria have superoxide dismutase (SODM), which converts superoxide radicals into hydrogen peroxide and oxygen (see Fig. C). Aerobic and most facultatively anaerobic bacteria have catalase, which converts hydrogen peroxide to water and oxygen (see Fig. C). Many oxygen-tolerant anaerobic bacteria have a peroxidase, which converts hydrogen peroxide to water by utilizing NADH2 (see Fig. C; click on it to enlarge it).
Updated: 2017-11-09.
Anaerobic cultivation
Anaerob incubation by commercial gas-generating envelopes in sealed anaerobic jar. An indicator strip that turns white in anaerobic atmosphere is included in the jar. - Click on the image to enlarge it.
Introduction
Some pathogenic bacteria are anaeribic and have to be cultivated in an oxygen-free atmosphere. The method of choice is dependent upon how anaerobic the bacterium to be cultivated is. There are strictly anaerobic bacteria, which cannot withstand oxygen at all, and there are aerololerant anaerobic bacteria, which will survive if the exposure to oxygen is not too long.
Strictly anaerobic bacteria
If you work with strict anaerobes, or if you have large amounts of samples to cultivate from, you may prefer a so-called anaerobic chamber where you work with rubber gloves sealed to the chamber. The anaerobic chamber maintains a constant temperature and the atmosphere may e.g. consist of nitrogen (N2) and carbon dioxide (CO2).
Aerotolerant bacteria
If you work with aerotolerant anaerobes maybe you prefer a so-called anaerobic jar where chemicals are used to absorb oxygen (O2) and generates CO2. There are commercial systems for this purpose. The anaerobic jar can be placed in thermostatic cabinet during the incubation.
Updated: 2020-02-26.
Antibiotics
Introduction
An antibiotic (pl. antibiotics) is a substance that selectively kills (bactericidal) or inhibits (bacteriostatic) growth of bacteria. Below is a compilation of various antibiotics, which are used in veterinary medicine. However, not all antibiotics mentioned below are necessaily registered for use on animals in Sweden or other countries. As for treatments of specific diseases in different species we refer to The Guidelines of the Swedish Veterinary Association (SVF). There you can find all the general guidelines for antibiotic treatment of animals and specific guidelines for small animals, horses and farm animals.
Bacteria may be resistant to some antibiotics and the resistance may be natural or acquired. Natural resistance means that a bacterium is resistant to an antibiotic due to its natural capacity. Mycoplasmas are e.g. naturally resistant to antibiotics which inhibit cell wall synthesis, because they have no cell wall. Acquired resistance means that a bacterium that was initially sensitive to a certain antibiotic, have developed resistance (by selection) because it has been exposed to this antibiotic. Antibiotic resistance is a very large and, unfortunately, growing problem in all health care because prescription of antibiotics has been too liberal in the past, and this is still the case in some countries, even within the EU.
From the National Veterinary Institut's (SVA's) website you can download the so-called SVARM- and SVARM/SWEDRES-reports, which are published annually by the Public Health Agency and SVA. The SVARM and SVARM/SWEDRES-reports provide a summary of antibiotic consumption and the resistance situation in Sweden in both veterinary and human medicine. The ability of bacteria to survive in the presence of an antibiotic can be tested by so-called susceptibility testing. The result of a susceptibility test is expressed as a MIC (Minimum Inhibitory Concentration) value, which has the unit mg/l (= µg/ml). In order to facilitate the interpretation of a MIC value, bacterial isolates are usually classified as sensitive (S), intermediate (I) or resistant (R) depending on the MIC value in relation to the "normal" MIC distribution of the corresponding bacterial species (see below).
Antibiotics are usually divided into different groups depending on their modes of action and chemical structure. Antibiotics, which are mentioned in the SVFs policy document appear in bold when they are first mentioned in the text below.
I. Inhibitors of cell wall synthesis.
A. Beta-lactams, where the so-called beta-lactam ring is included in the structure.
- Penicillins (e.g. aminopenicillin amoxicillin, ampicillin, benzylpenicillin, phenoxymethylpenicillin, cloxacillin and methicillin).
- Cephalosporins (e.g. cephalexin, cephalothin, cefotaxime and cefepime). Cephalosporins should be used very restrictively in the treatment of animals as they are also used to treat life threatening diseases in humans. Use of the third and fourth generation of cephalosporins is regulated in the Swedish Constitution SJVFS 2013: 42, Saknr D9.
- Cefamycins (latamoxef) is similar to the cephalosporins.
- Carbapenems (e.g. imipenem, meropenem and aztreonam) are currently banned from use in the treatment of animals as they are also used for the treatment of life-threatening diseases in humans (Swedish Constitution SJVFS 2013: 42, Saknr D9, Annex).
B. Other inhibitors of cell wall synthesis.
- Glycopeptides (e.g. vancomycin and teicoplanin) block the transpeptidation by binding to -D-alanine-D-alanine- in the side peptide of the peptidoglycan. Glycopeptides are today prohibited to use for the treatment of animals, as they are also used for the treatment of life-threatening diseases in humans (Swedish Constitution SJVFS 2013: 42, Saknr D9, Appendix).
Examples of resistance mechanisms:
A transpeptidase is involved in the synthesis of the cell wall of bacteria. This transpeptidase binds to the dipeptide -ala-ala- and links peptides to be included in the cell wall, into a network. The beta-lactam ring is a structural analogue of -ala-ala- and can, therefore, compete with ala-ala which means that a stable cell wall cannot be formed. Mutations in the transpeptidase or other so-called penicillin binding proteins (PBP) may lead to penicillin no longer binding and, therefore, the bacterium becomes resistant.
Some bacteria are naturally possessing a chromosomal or plasmid encoded penicillinase, i.e. an enzyme that hydrolyzes the beta-lactam ring. In the presence of beta-lactams, these bacteria may be stimulated to increase production of beta-lactamase and if the beta-lactamase gene is plasmid-encoded, it can be transferred to other bacteria.
Clavulanic acid, which is a structural analogue of penicillins, can inhibit penicillinases by blocking their active centers. Clavulanic acid is particularly active against plasmid born beta-lactamases and is used therapeutically. Clavulanic acid is not an antibiotic in itself, since it only works in combination with certain beta-lactams.
II. Inhibitors of nucleic acid synthesis.
A. The fluoroquinolones (such as ciprofloxacin, difloxacin, enrofloxacin, ibafloxacin, marbofloxacin and orbifloxacin) inhibit the enzymes DNA gyrase and topoisomerase, which are needed for DNA replication. Fluoroquinolones must be used very restrictive for the treatment of animals because they are also used for the treatment of life-threatening diseases in humans. Use of the third and the fourth generation of fluoroquinolones are regulated in the Constitution SJVFS 2013: 42, Saknr D9.
B. Rifampicin inhibits DNA-dependent RNA polymerase, which in turn blocks the protein synthesis since the synthesis of the mRNA is inhibited. Rifampicin must only be used for the treatment of Rhodoccus-infectioner (Constitutional SJVFS 2013: 42, Saknr D9, Annex).
C. Nitroimidazoles (e.g. dimetridazole, metronidazole and ronidazole) reacts with nucleoproteins under anaerobic conditions, which in turn results in breakage of the DNA helix. Metronidazole is one of the few antibiotics that are effective against Clostridium difficile-induced diarrhea in humans and should, therefore, only be used for animals in exceptional cases.
Examples of mechanisms of resistance:
Resistance to fluoroquinolones arise by mutation(s) in the genes for the DNA gyrase and/or topoisomerase, which makes the fluoroquinolones unable to block these enzymes.
III. Inhibitors of protein synthesis.
A. Aminoglycosides (e.g., gentamicin, neomycin, streptomycin and dihydrostreptomycin) bind to the 30S subunit of the bacterial ribosomes and cause misreading of the genetic code.
B. Tetracyclines (chlortetracycline, oxytetracycline and doxycycline) interfere with the aminoacylated tRNA chains and prevents binding of amino acids to the growing polypeptide chain. This effect leads in turn to inhibition of protein synthesis.
C. Chloramphenicol (chloromycetin) inhibits the enzyme peptidyl transferase, linking amino acids to the growing polypeptide chain. This inhibition results in blocking of protein synthesis.
D. Macrolides (e.g. tylosin and erythromycin) and lincosamides (e.g. lincomycin and clindamycin) binds to the 23S rRNA of the bacterial ribosome and prevents translocation along the mRNA chain. Ketolides belong to the group of macrolides, but they have a broader bacterial spectrum. Azalides (e.g. azithromycin) and streptogramins also belong to the group of macrolides.
E. Pleuromutilins (e.g. valnemulin and tiamulin) bind to the peptidyl transferase of the 50S subunit, thereby preventing amino acids from connecting with the growing polypeptide chain.
F. Fusidic acid is a steroid derivative, which interferes with the release of elongation factor EF-G when amino acids are coupled to the growing polypeptide chain.
G. Oxazolidinones bind to the 23S rRNA of the bacterial ribosome preventing translocation along the mRNA chain. Oxazolidinones are today prohibited to be used for the treatment of animals, as they are also used for the treatment of life-threatening diseases in humans (constitution SJVFS 2013: 42, Saknr D9, Appendix).
Examples of mechanisms of resistance:
Resistance to aminoglycoside may arise by mutations in the ribosomal proteins, so that the macrolide can no longer bind to the ribosome. Some bacteria carry plasmids encoding aminoglycoside modifying enzymes. Such enzymes inactivate aminoglycosides and can easily spread in a bacterial population.
Lincosamide and macrolide resistance can occur by bacterial methylation of a specific nucleotide in the 23S rRNA or by mutations in the 23S rRNA gene, which result in that the macrolide will no longer bind to the ribosome.
IV. Inhibitors of functions in the cell membrane.
A. Polypeptide antibiotics (such as polymyxin B and colistin) affect cell membrane permeability by binding to LPS, and interfering with phospholipids. This allows the essential metabolites to leak out from the bacterial cell.
B. Cyclic lipopeptides (such as daptomycin) bind to the bacterial cell membrane, which is then depolarized. This leads to inhibition of DNA, RNA and protein synthesis.
V. Inhibitors of different steps in the metabolism.
A. Folic acid antagonists
- Sulfonamides (eg, sulfa and sulfadoxine) inhibits the synthesis of folic acid by competing with the structural analog para-aminobenzoic acid. Folic acid is a coenzyme required for the synthesis of certain amino acids and nucleotides and which eukaryotic cells can take up from the environment.
- Trimethoprim inhibits the enzyme dihydrofolate reductase. Sulfa and trimethoprim are often combined to counteract the emergence of resistance.
B. Furantoins (such as nitrofurantoin and furadantin) are transformed by bacterial flavoproteins to reactive intermediates, which inhibit bacterial energy metabolism as well as the synthesis of DNA, RNA, proteins and the cell wall. Furantoins affect so many different steps in of bacterial metabolism and, therefore, resistance does not easily arise. Consequently, furantoins are used against urinary tract infections in primarily dogs to reduce the use of aminopenicillins, against which resistance is more easily incurred. Resistance against furantoins occurs, but is uncommon.
Examples of mechanisms of resistance:
Point mutations in the genes for some of the enzymes involved in the synthesis of folic acid may lead to resistance because these modified enzymes are no longer blocked. Enterococci are naturally resistant to sulfonamides because they absorb folic acid from the environment.
VI. General mechanisms of resistance:
By decreased permeability or increased efflux, bacteria can develop resistance mechanisms, which are effective against different types of antibiotics. Decreased uptake may be due to mutations in porins and increased efflux may be due to mutations in the so-called efflux pumps. Efflux pumps are protein complexes which can pump the drugs out of the cell by active transport. Most often, the natural substrates of efflux pumps are bile salts, adhesins, toxins and other proteins.
VII. Abbreviations
ESBL, means "Extended-Spectrum Beta-Lactamases", which implies that the bacterium has a beta-lactamase with extended spectrum (can hydrolyse different kinds of beta-lactams). ESBL is sometimes used, a bit sloppy, for a group of multi-resistant bacteria, but is actually a name given to a group of enzymes, which are present in some multi-resistant bacteria. ESBL confer resistance to penicillins and many cephalosporins. ESBL are often plasmid encoded and may be transmitted between gram negative bacteria.
ESBL-CARBA (or ESBLCARBA), is the term for a mechanism of resistance in enteric bacteria, which causes resistance to most penicillins, cephalosporins and carbapenems. In Sweden, findings of ESBLCARBA in animals are notifiable to the County Administrative Board and the Board of Agriculture.
MRS means Methicillin Resistant Staphylococci. MRS is a designation for staphylococci in general, which is resistant to all penicillins (such as methicillin) and cephalosporins. In Sweden, findings of MRS in animals are notifiable to the County Administrative Board and the Board of Agriculture.
MRSA means Methicillin Resistant Staphylococcus Aureus. MRSA is the designation of Staphylococcus aureus strains, which are resistant to all penicillins (such as methicillin) and cephalosporins. In Sweden, findings of MRSA in animals are notifiable to the County Administrative Board and the Board of Agriculture.
MRSP means Methicillin Resistant Staphylococcus Pseudintermedius. MRSP is the designation of Staphylococcus pseudintermedius strains, which are resistant to all penicillins (such as methicillin) and cephalosporins. In Sweden, findings of MRSP in animals are notifiable to the County Administrative Board and the Board of Agriculture.
VRE means Vancomycin Resistant Enterococci.
VIII. More information
More information about the antibiotic resistance situation and antibiotic consumption in various European countries (including Sweden) can be obtained on the website of the European Centre for Disease Prevention and Control when clicking on "Health Topics" and then select "Health Topics A-Z" and then "Antimicrobial resistance". There you can select e.g., "Antimicrobial resistance interactive database (EARS-Net)" under Interactive Databases. Another important source of information on antibiotic susceptibility in the form of MIC values is the website EUCAST (European Committee on Antimicrobial Susceptibility Testing), where you can search a database for a specific antibiotic or a particular bacterium.
See also Susceptibility Testing on this website.
Updated: 2025-06-25.
Aseptic - Antiseptic
To work aseptically means that you work in a way that will prevent disease-causing microorganisms to contaminate the material you are working with, without the use of chemical disinfectants. Chemical disinfectants (= germicides) are substances used to inhibit the growth of or to kill microorganisms on either an object or a body surface.
An antisepticum is a substance that is used to inhibit the growth of or to kill microorganisms on a body surface.
A bactericide is a substance that kills bacteria. A bacteriostatic agent (= bacteriostat) is a substance that inhibits the growth of bacteria.
A germicide is a substance that kills not only bacteria but also other microorganisms.
Updated: 2014-01-22.
Bacteriophages
Introduction Viruses are organisms which parasitize the host cell's protein synthesis machinery. Bacteriophages (phages or bacterial viruses) are viruses that infect and replicate in bacterial cells. Phages may have dsDNA, ssDNA, dsRNA or ssRNA as chromosome and the chromosome can be circular or linear. Phages have either a lytic or a lysogenic cell cycle.Lytic phages
Lytic phages injects their chromosome into the bacterial host. To be able to do this, the phage has to adhere to specific receptors on the surface of the host bacterium. Then, the chromosome is replicated and phage proteins are synthesized by means of the host bacterial ribosome etc. When new phage progeny has formed, the bacterial host cell will lyse and the phage particles will find new bacterial host cells.Lysogenic phages
Lysogenic (= temperate) phages does not lyse the host bacterium immediately, but its chromosome can instead be integrated into the bacterial genome and there it exist as a so-called prophage (endogenous phage). The prophage will then replicate during the bacterial cell division and is passed on to successive generations. The phage will exist as a prophage until the environment for the bacteria deteriorate. Then the prophage become active, form new phage particles and finaly lyse the host cell.Lysogenic conversion
Some bacteria (e.g. Corynebacterium diphtheria, Clostridium botulinum, Shigella dysenteriae, Escherichia coli type VTEC and Streptococcus pyogenes) are pathogenic only if they carry a prophage. They are then said to have undergone lysogenic conversion and it is thus the genes of the prophage that encode important virulence factors.Phages as diagnostic tools
Phages are not only species-specific, but in many cases also strain-specific and therefore, they can be used for subtyping of bacteria. Phagetyping is used for epidemiological studies of Salmonella enterica subsp. enterica. Phagetyping is perfomed by testing as a set of phage types and examine which of these can lyse the current salmonella strain. To perform the testing, a drop of bacteria is spread onto an agar plate, which is then allowed to dry on the surface. Then, small drops of phages with defined specificities are added. After incubation, the agar plate is inspected for plaques, which are formed where phages for which the bacterium is sensitive, have been applied. Exemples of other bacterial genera for which phage typing is used: Bacillus, Campylobacter, Clostridium and Staphylococcus.Phage therapy
Since the problem of multi-drug resistant strains of bacteria increases worldwide, the interest in treating bacterial infectious diseases by phage therapy has increased. For this purpose, lytic phages are best suited. The method is still in the experimental stage, but many researchers believe that it has great potential in both human and veterinary medicine.Updated: 2013-03-03.
Biofilm and Quorum Sensing
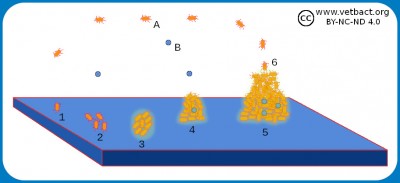
The figure shows a schematic picture of how a biofilm is formed. Two different bacterial species (A and B) are first planktonic. One of the bacteria (1), which has adhesion ability, adheres to the surface and begins to multiply there (2). After a while, a monolayer has been formed and polysaccharides (glycocalyx) are secreted (3). Then the monolayer begins to grow in height and a colony, which is encapsulated in glycocalix, is formed (4). This is the beginning of a biofilm, which may contain several different bacterial species and also fungi (yeast or mould). When the biofilm has grown to a certain size, bacteria (6) or clumps of bacteria are released, which can attach to new places and form more biofilm.
Image: Karl-Erik Johansson (BVF, SLU) - Click on the image to enlarge it.
Bacteria that grow freely in liquid media are said to be planktonic, but some bacteria can also grow on solid substrates, forming a so-called biofilm. Biofilms can exist on live or dead surfaces and occur in nature, on materials in industrial processes and in different health care situations. Biofilms consist of population(s) of bacteria, which adhere to a surface and to each other and are enclosed in a network (matrix) of biopolymers. The formation of a biofilm starts with bacteria that adhere to a surface by means of e.g. fimbriae (pili) where they bind irreversibly and initially grow as a monolayer. Then they form several layers and start to produce some kind of a biopolymer (extracellular matrix) that often consists of the same material as the capsule, but in a looser structure. The biopolymer is made up of polysaccharides and is called glycocalyx (capsule). Dextran is one example of such a polysaccharide. Biofilms consist of one or more bacterial populations (species), glycocalyx, DNA and proteins.
A bacterial species that can not itself adhere to a surface, can often become attached to pre-existing bacterial glycocalyx and grow as a biofilm. Bacteria in biofilms are more resistant to antibiotics, detergents and phagocytosis than planktonic bacteria. Bacteria (Streptococcus spp.) can grow as biofilms on teeth (= plaques), on implants (e.g. heart valves), in plastic tubes, which transport nutrients etc. In patients with cystic fibrosis Pseudomonas aeruginosa may grow as a biofilm in the lungs. In biofilms, bacteria can communicate with each other by means of chemical signal substances (quorum sensing) to control gene expression in the whole population.
Link to a video montage on YouTube which is recommended:
Biofilm: "What Are Bacterial Biofilms?"
Quorum sensing: "Quorum sensing, bacteria talks"
Updated: 2021-01-28.
Capsule
Introduction
Many bacteria have a so-called capsule, which usually consists of polysaccharides and sometimes also of other substanances. The capsule is a structure outside the bacterial cell envelope and it can also be defined as the outer cell envelope of the bacterium. The capsule is a well-defined layer that can constitute a virulence factor for both gram-positive and gram-negative bacteria. If the amorphous and viscous secretion, which constitutes the capsule, diffuses into the surroundings and remains as loosely undefined layer without a clear outer boundary, one use instead to speak of a slime layer. In summary, capsules and slime layers are usually called glycocalyx. A biofilm is formed when many bacteria are enclosed in a common slime layer.
Composition
The polysaccharide capsule of a bacterium can consist of many different linear polysaccharides. These polysaccharides are made up of repeating subunits, which consist of one to six monosaccharides. The diversity is enormous and it has been shown that only Escherichia coli can produce about 200 different types of polysaccharides. The capsule may also consist of peptidoglycan (as the cell wall), poly-muramic acid (one of the cell wall components), glycoproteins and poly-D-glutamic acid as in Bacillus anthracis. To be able to see the capsule in a microscope, one can use a special capsule staining method with methylene blue and this has been utilized for B. anthracis (see the bacterial page).
Function
The capsule is considered a pathogenicity factor, as it can increase the ability of bacteria to cause disease. The capsule protects the bacterium from phagocytosis of macrophages. The capsule contains water and, therefore, protects bacteria from dehydration. The capsule increases the ability of bacteria to bind to surfaces and to each other. When bacteria form a biofilm, they are less susceptible to antibiotics and some other bacteriostatic agents. The capsule can also protect bacteria from attack by certain bacteriofages.
Updated: 2020-04-09.
Carbohydrates
Introduction
Carbohydratesis a very important source of energy for bacteria and the name carbohydrate indicates that they are substances that contain carbon (C), as well as hydrogen (H) and oxygen (O) in the same proportions as in water (there are, however, some exceptions). Bacteria, however, cannot utilize di-, tri- or polysaccharides for glycolysis unless they have enzyme systems that can degrade them into monosaccharides. Examples of carbohydrates, which bacteria can metabolize by fermentation to produce an acid, is provided below.
Identification of bacteria
Fermentation may be detected by adding a pH indicator to the culture medium (liquid or solid) and this can be utilized for the differentiation of bacteria.
Different laboratories are using different combinations of carbohydrates in liquid media and, and the selection of combination. is depending on the suspected bacterium. Carbohydrates that are used for identification of bacteria are listed below. Sometimes you want to perform fermentation tests on agar plates, since it gives more reliable results in some cases.
- A.1. Aldotriose monosaccharides:
The aldotriose monosaccharide has thee carbon atoms and one aldehyde group in position 1. - Glyceraldehyde, which is the simpliest one of all aldoses (formula: C3H6O3).
- A.2. Triose monosaccharide alcohol:
In a triose monosaccharide alcohol, the carbonyl group (=O) has been reduced to a hydroxyl group (-OH). - Glycerol, which is the simpliest one of all saccharide alkohols (formula: C3H5(OH)3).
- B.1. Aldopentose monosaccharides:
Aldopentose monosaccharides have five carbon atoms and one aldehyde group in position 1. - L-arabinose
- D-ribose
- D-xylose
B.2. Ketopentose monosaccharides:
Ketopentose monosaccharides have five carbon atoms and one carbonyl group in position 2. - D-ribulose
- B.3. Pentose monosaccharide alcohols:
In a pentose monosaccharide alcohol, the carbonyl group (=O) has been reduced to a hydroxyl group (-OH). - adonitol (= ribitol)
- xylitol
- C.1. Aldohexose monosaccharides:
Aldohexose monosaccharides have six carbon atoms and one aldehyde group in position 1. - galactose
- glucose
- mannose
- L-rhamnose (= 6-deoxy-L-mannose)
- fucose, is an aldodeoxyhexose, which lacks the hydroxyl group in position 6.
- C.2. Ketohexose monosaccharides:
Ketohexose monosaccharides have six carbon atoms and one carbonyl group in position 2. - fructose
- sorbose
- C.3. Hexose monosaccharide alcohols:
In hexose monosaccharide alcohols has the carbonyl group (O=) been reduced to a hydroxyl group (OH-). - dulcitol
- inositol, is not a classical sugar, but a carbohydrate in the form of a av sixfold alcohol of cyklohexan (formula: C6H12O6)
- mannitol
- sorbitol
- D. Hexose disaccharides:
Hexose disaccharides are composed of two hexose monosaccharides linked to each other by a glycosidic bond. - cellobiose, is composed of two D-glucose moieties linked to each other by a β-(1→4) glycosidic bond
- lactose, is composed of one D-galactose and one D-glucose moiety linked to each other by a β-(1→4) glycosidic bond
- maltose, is composed of two D-glucose moieties linked to each other by a α-(1→4) glycosidic bond
- melibiose, is composed of one D-galactose and one D-glucose moiety linked to each other by a α(1→6) glycosidic bond
- sucrose (= saccharose or cane sugar), is composed of one D-glucose and one D-fructose moiety linked to each other by a (2→1) glycosidic bond
- trehalos, is composed of two D-glucose moieties linked to each other by a (1→1) glycosidic bond
- E. Hexose trisaccharides:
Hexose trisaccharides are composed of three hexose monosaccharides linked to each other by glycosidic bonds. - raffinose, is composed of one D-galactose, one D-fructose and one D-glucose moiety
- melezitos, is composed of av two D-glucose and one D-fructose moiety
- F. Polysaccharides:
Polysackarider are composed of long linear or branched carbohydrate chains, where the monosaccharide moieties are linked together by glycosidic bonds. - glycogen, is primarily composed of D-glucose moieties
- inulin, is composed of D-fructose and a terminal D-glucose moiety
- starch, is composed of linear and branched D-glucose chains
- dextrin, is a mixture of polymers of D-glucose moieties linked to each other by α-(1,4) or α-(1,6) glycosidic bonds
- G. Glycosides:
Glycosides are composed of a carbohydrate(s), which is linked to another molecule of non-carbohydrate nature, by a glycosidic bond. - salicin, is an alcoholic β-glycoside of o-hydroxy benzyl alcohol containing one glucose moiety
- amygdalin, is a disaccharide, which is composed of two glucose moieties, one benzene ring and a cyano group (-CN).
Updated: 2019-02-24.
Cell wall and cell membrane
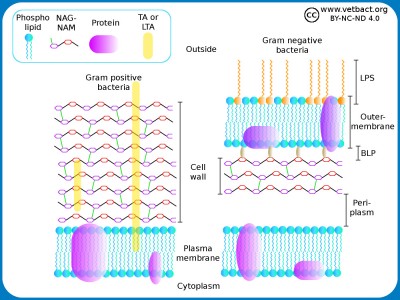
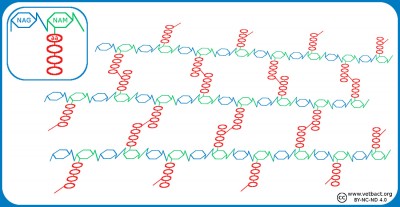
Schematic illustration of the cell envelope of gram positive and gram negative bacteria. NAG-NAM stands for N-acetylglucosamine and N-acetylmuramic acid, which are the building blocks of the cell wall. TA and LTA stand for teichoic acid and lipoteicoic acid, respectively. These components stabilize the cell wall and anchors it in the plasma membrane of gram positive bacteria. LPS stands for lipopolysaccharide and BLP stands for Brown's lipoprotein, which binds the cell wall to the outer membrane of gram negative bacteria.
Image: Karl-Erik Johansson (BVF, SLU). - Click on the image to enlarge it.
Introduction
The cell wall and the cell membrane are very important structures of bacteria because they form the bacterial contact surface with the environment. For bacteria, the cell wall together with the cell membrane and also the outer membrane (if present), for the cell envelope. Differences in cell envelope structures can be used to differentiate different groups of bacteria e.g. by gram staining. For some antibiotics, the fact that there is a large difference between the bacterial cell envelope and the cell envelope of higher organisms (eukaryotic cells) is exploited. There are two different main types of structures in the cell envelopes of bacteria and since it can be utilised in gram staining, these bacteria are said to be gram positive or gram negative. The differences between the gram positive and the gram negative cell envelopes are shown in the very simplified in figure.
The components of the cell envelope
In gram positive bacteria, the cell wall is 20-80 nm thick, while in gram negative bacteria it is only 5-10 nm thick. The cell wall consists of peptidoglycan, which is made up of repeating units of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). The peptidoglycan chains are cross-linked to each other through peptide bridges and form a continuous network around the bacterium. This network can be considered a giant molecule. Gram-negative bacteria also have an outer membrane containing lipopolysacharide (LPS) to protect against the environment. Both the plasma membrane and the outer membrane are about 8 nm thick and contain proteins that are important for the bacterial functions. Some authors include both peptidoglycan and outer membrane of gram negative bacteria in the term cell wall. The periplasm is the space that exists between the cell wall and the plasma membrane in gram negative bacteria. Some authors define the periplasm as the space between the plasma membrane and the outer membrane. The periplasm is gelatinous and contains enzymes which are important for the metabolism of the bacteria.
In addition to the above components, bacteria may have flagella, fimbriae and capsules on the cell surface (not shown in the figure). In the lecture on chlamydia, it is stated the chlamydia lack the peptidoglycan, which is not correct according to current knowledge.
The functions of the cell envelope
The cell envelope is, of course, vital for the bacterium, since its surroundings can be very harsh and thus the envelope provides protection for the bacterium. There are various enzyme systems that are bound to the plasma membrane, e.g. those that are part of the elektron transport chain and some of the enzymes that participate in the synthesis of the cell wall. The molecular motor, which consists of various protein subunits and drives the movement of flagella, is anchored in the plasma membrane of the bacterium. Many bacteria have so-called secretion systems, which are of great importance for their pathogenic ability. The secretion systems also consist of a number of different membrane-bound protein subunits. Porins are membrane-bound proteins that can transport substances through the plasma membrane and the outer membrane if present.
Exceptions
There are bacteria that have a completely different structure of the cell envelope than described in the figures above.
Fylum Mycoplasmatota
Bacteria belonging to the phylum Mycoplasmatota, which includes the organisms commonly known as mycoplasmas, are lacking a cell wall because they are unable to produce the peptidoglycan. These bacteria usually live in physiological environments where the mechanical protection of the cell against osmotic lysis is not needed. More information about mycoplasmas can be found in a series of lectures.
The Chlamydiaceae family
It was long believed that bacteria within the Chlamydiaceae family have no peptidoglycan, but instead cysteine-rich proteins, which stabilize the outer membrane, through disulfide bonds. However, a few years ago, a research group showed that the reticular bodies of chlamydia have a thin peptidoclycan, which is important in cell division, which in chlamydia occurs in host cell inclusions. In the lecture on chlamydia, it is stated that chlamydias lack the peptidoclycan, which is not correct according to current knowledge,
Order Corynebacteriales
The order Corynebacteriales, to which the families Corynebacteriaceae, Mycobacteriaceae and Nocardiaceae belong, have a cell envelope, most similar to that of gram-positive bacteria. These bacteria thus have a plasma membrane, peptidoglycan and outside the peptidoglycan a polysaccharide layer (arabinogalactan) as well as mycolic acid, which with its hydrocarbon chains binds to a single layer of lipids. Mycobacteria are said to be acid-resistant because they have an unusually stable cell envelope.
Updated: 2024-03-20.
Coliform bacteria
Coliform bacteria (coliforms) are Gram-negative, non-spore forming and rod-shaped bacteria, which are also facultatively anaerobic, lactose-positive (during formation of gas) and oxidase-negative when grown for 24-48 h at 37°C. Heat tolerant coliforms are bacteria that also produce gas at a temperature of 44.0°C during cultivation for 24 h.Coliform bacteria are used in food production as indicator bacteria for general hygiene. For microbiological analysis of water, coliforms are always used as indicator bacteria.
Examples of coliforms: Escherichia spp. (including E. coli), Citrobacter spp. Enterobacter spp., Hafnia spp. and Klebsiella spp.
Updated: 2015-06-17.
Colony Forming Unit (CFU)
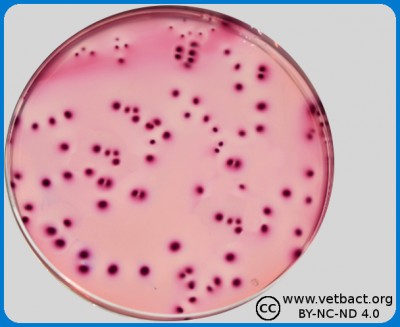
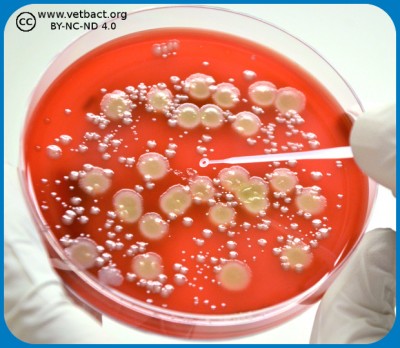
Colony Forming Units (CFU) could be quantified by pour plate technique, in this figure bacteria belonging to Family Enterobactericaeae in VRBG from a food sample. - Click on the image to enlarge it.
The amount of bacteria in a solid medium, for example, food is often expressed as colony forming units per gram, (CFU/g), whereas the number of bacteria in a liquid sample is expressed as colony forming units per ml (CFU/ml). This value can be determined by first making a tenfold dilution series of the sample. Then, a known volume (usually 1 ml or 0.1 ml) is taken from each dilution and is applied to appropriate agar plates. After incubation, the number of colonies is counted on the agar plate with 15-150 colonies, which is considered to give the most accurate result. When calculating the actual number of bacteria in the sample, the dilution factor also has to be considered.
In assessing urine samples, the number of bacteria per ml (CFU/ml) is of great importance. Urine is collected with a loop of a known volume (1 µl) and distributed on the agar plate to facilitate counting of the number of colonies. The amount of bacteria in urine is usually classified in the following way.
-
0 cfu/ml: no growth
-
<25 000 cfu/ml: sparse growth, which is regarded as normal (contamination) flora
-
25 000 - 100 000 cfu/ml: moderate growth
-
>100 000 cfu/ml: rich growth
Note that this method to determine the amout of bacteria in urine, only gives a rough estimate of the actual number.
Bacteriuria is defined as the presence of bacteria in urine in almost pure culture with a concentration of more than 100 000 CFU/ml. A patient is said to have urinary tract infection (UTI) when there is bacteriuria and clinical symptoms of infection.
The reason for using the term CFU is that it is not certain that each colony originates from a single bacterial cell, because some bacterial species easily form aggregates in suspension cultures. Only living (or rather culturable) bacteria will give rise to colonies.
Updated: 2020-02-20.
Counting bacteria
Fig. 1. In surface spreading 0.1 ml is distributed with a sterile spreader on the solid agar in this case mCCDA [Image: Moa Skarin & Ingrid Hansson (BVF, SLU)]. - Click on the image to enlarge it.
Bacteria can be counted in different ways, and here are three fundamentally different methods:
- Determination of CFU (Colony Forming Unit). Samples that should be analysed are diluted and spread on/in agar plates, which are incubated under different conditions depending on the method for analysis, i.e. which bacteria could be expected from the current issue. This method is relatively accurate, provided that the dilution is performed in a proper manner and that the bacteria do not form aggregates in suspension cultures. Disadvantage: the answer is obtained after at least 24 h depending upon which agar is used and which bacterium you look for. See also Colony Forming Unit.
Depending upon test parameter, surface spreading or pour plate technique is used in food microbiology. A ten-fold serial dilution is prepared and for surface spreading, 0.1 ml is used from the different dilutions and 1.0 ml is used for pour plating. In surface spreading 0.1 ml from the dilution is distributed by a sterile spreader on the whole agar surface (see Fig. 1). However, in pour plating, the bacteria are directly mixed in the whole volume of the "melted" agar in the petri dish (see Fig. 2) and not only on the surface. The melted agar has a temperatur of 45°C, which does not kill the bacteria, but keeps the agar fluid.
- Spectrophotometric determination of light scattering. The method provides a relative measure of the number of bacteria, but can be calibrated with a standard curve. See Growth Curve.
- Direct counting in a so called Bürkner chamber that can be placed under a microscope. The method is relatively uncertain and is not used that often.
Updated: 2020-02-13.
Direct smear
Direct smear is perfermed by making a suspension of a clinical sample and smear it out directly onto a microscope slide without first making a bacterial cultivation. Then the sample is fixed and stained by simple stain techniques or differential stain techniques, to provide an indication of whether the sample contains pathogenic bacteria.Updated: 2013-02-23.
Disease types
Introduction
In both veterinary and human medicine, infectious diseases (caused by bacteria, fungi, parasites or viruses etc.) are usually characterized according to different criteria, and here are some examples of classification of diseases within these two disciplines.
Enzootic corresponds to endemic in human medicine and an enzootic disease is a disease, which is always present in a certain animal population, but which at a certain time only affects a small number of animals. Enzootic pneumonia in pigs, caused by Mycoplasma hyopneuminiae, is an example of a bacterial-induced enzootia.
Epizootic corresponds to epidemic in human medicine and an epizootic disease, is a serious animal infectious disease that is widespread. Anthrax, which is caused by Bacillus anthracis, is an example of a bacterial epizootic disease.
A panzootic is a serious infectious disease, which spreads over large parts of the world and affects one or several species in many countries. African swine fever is an exaple of a virus cause panzootic.The equivalent term in human medicin is pandemic and cholera, caused by the bacterium Vibrio cholerae, gave rise to three pandemics in the 19th century.
Zoonosis is a term used in both veterinary and human medicine, as it refers to infectious diseases, which can be transmitted between animals and humans via food, direct contact with infectious animals or through indirect contact with infectious animals via e.g. insect bites. Brucellosis, campylobacteriosis, bovine tuberculosis and salmonellosis are examples of zoonoses, which are caused by the following zoonotic bacteria: Brucella melitensis, Campylobacter jejuni, Mycobacterium bovis and Salmonella enterica subsp. enterica.
Updated: 2024-02-08.
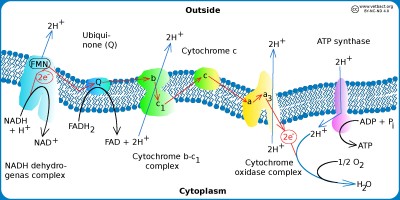
Electron transport chain
The figure shows a schematic view of the electron transpor chain in bacteria with aerobic metabolism. Hydrogen ions (protons) are pumped out (blue arrows) out of the cell by using three different enzyme complexes and a proton gradient is formed over the plasma membrane. The proton gradient is the energy (proton motive force) used by the ATP synthase to produce ATP. The electrons jump gradually between the various components of the enzyme complexes, which have increased electronegativity from left to right in the figure. The higher the electronegativity, the stronger the attraction of electrons. Oxygen is the component of the system that has the highest electronegativite and it is also the final electron acceptor.
Image: Karl-Erik Johansson (BVF, SLU) - Click on the image to enlarge it.
Introduction
During the glycolysis, two molecules of pyruvate, each having 3 carbon atoms, are formed from a glucose molecule having 6 carbon atoms. Under aerobic conditions and if the current bacterium has aerobic metabolism, the pyruvate molecule is not fermented, but can instead be converted to acetyl-Coenzyme A (actyl-CoA). The acetyl group in acetyl-CoA has two carbon atoms since carbon dioxide (CO2) has been cleaved off and can now enter the Krebs cycle (= citric acid cycle), as an acetyl group. The Krebs cycle is described in a separate section and is a precursor to the electron transport chain (= respiration chain).
The electron transport chain
All enzymes required for the electron transport chain of bacteria are membrane bound as in eukaryotic cells, but in bacteria these molecules are present in the plasma membrane because bacteria have no mitochondria. The hydrogen ion gradient, which drives ATP synthesis, is thus generated across the plasma membrane. The electron transport chain consists of a series of enzyme complexes, which incrementally take care of the electrons that are formed when NADH and FADH2 (from the Krebs cycle) are oxidized to NAD+ and and FAD, respectively. At the same time, hydrogen ions (protons) are pumped out of the bacterial cell. When these hydrogen ions then pass to another membrane-bound enzyme complex (ATP synthase) on their way back into the cytoplasm, ATP is generated while the electrons are finally taken care of by oxygen and water is formed with the hydrogen ions present in the cytoplasm.
Variants of the electron transport chain
There are various variants of the electron transport chain and Escherichia coli, for instance, lacks cytochrome C oxidase as do most other bacteria in the family Enterobacteriaceae. These bacteria have instead a terminal cytochrome bo3 oxidase. Some bacteria have enzyme systems for anaerobic respiration, where molecules other than oxygen form the terminal electron acceptor. The following molecules may be used: nitrate, nitrite, ferric iron (Fe3+), sulfate, carbon dioxide, and small organic molecules such as fumarate. In anaerobic respiration, we talk about terminal reductases instead of terminal oxidases.
Updated: 2021-10-06.
Flagella and fimbriae
Bacteria can have different types of protrusions from the surface, known as flagella and fimbriae. Fimbriae are also called pili.
The primary mission of the flagellum is to provide the bacteria motility. They can then "swim" with the help of their flagella. Motility may be important for the ability of bacteria to cause disease and, therefore, flagella are regarded as a patogenicity factor. The flagellum is typically 10-30 nm in diameter and 5-15 µm in length. Bacteria can be classified by the number of flagella and how the flagella are arranged on the cell surface as follows:
- Monotrichous bacteria have a single flagellum (e.g. Vibrio cholerae), which is said to be polar.
- Lofotrichous bacteria have many flagella, extending from one or two opposing areas on the cell surface.
- Amfitrichous bacteria have flagella on each end of the cell.
- Peritrichous bacteria have flagella scattered all over the cell surface (eg, Escherichia coli).
- Atrichous bacteria are lacking flagella.
The flagellum is made up of three parts, consisting of different proteins:
- The basal body, consisting of a system of rings, which are anchored in the cell envelope. The inner rings (S and M) are the engine which drives flagellar movement.
- The hook, which sits near the cell surface and connects the engine with the long flagellar filamentet.
- The filament, consisting of many subunits of the protein flagellin. Flagellin molecules form a hollow tube through which the new flagellin molecules are transported when the tube is extended. Flagellin is antigenic and is the so-called H antigen.
Bacteria within the phylum Spirochaetota have so-called periplasmic flagella (= axial filaments = endoflagella), which are localized in the periplasmic space and gives these bacteria a very characteristic corkscrew like movement.
Fimbriae (= pili) is another type of hair-like projections which in some exceptional cases (type IV pili) can give bacteria motility (eg in Psueudomonas sp.), but above all contribute to bacterial adhesion. So-called F-pili and sex pili give bacteria the opportunity to exchange genetic material (DNA).
Updated: 2025-05-23.
Genome
Introduction
The term genome refers to the complete set of genetic material in a cell. Genome size is usually expressed in Mb or Mbp (= mega base pairs). Thus, 1 Mbp = 1 000 000 bp. The size of the bacterial genome varies between 0.5 and 10 Mbp. Bacterial genomes comprise the chromosome(s) and possibly also plasmid(s). Bacteria are haploid, unlike higher organisms, which are diploid. Gametes of higher organisms are, however, also haploid. The complete genome sequences have been determined for about 30 000 bacterial strains and there are approximately 20 000 published genome projects for bacteria, where the sequence fragments have not been aligned into one single sequence (a so-called contig) .
Chromosome
The chromosome represents the primary genetic material, which is essential for the bacterium. Bacteria in general have a circular chromosome, but there are exceptions (see below).
Plasmid
Plasmids (see also Genome above) are secondary genetic material, not always essential for the bacterium. Bacteria which carry plasmids generally have one or more circular plasmids, but there are also exceptions. Plasmids generally constitute up to 10% of the genome and they are replicated independently of the chromosome.
Exceptions
- Some species within the genera Brucella, Burkholderia, Leptospira och Vibrio have two cirkular chromosomes
- Species within the genus Borrelia have one linear chromosome (about 1 Mbp), which is relatively small.
- Species within the genus Borrelia also have linear plasmids and these are essential for the bacterium.
- Species within the genus Streptomyces have one linear chromosome, which is relatively big (about 10 Mbp).
Updated: 2020-09-23.
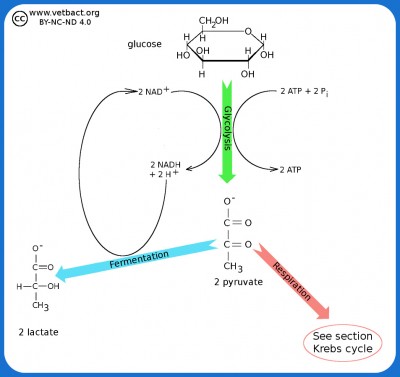
Glycolysis and Fermentation
The figure shows a very simplified picture of bacterial glycolysis and fermentation where the final product is lactate (the ionized form of lactic acid). The net gain of glycolysis is 2 ATP and 2 NADH per glucose molecule. However, the two molecules of NADH are used in the fermentation of pyruvate to lactate. Pyruvate acts as an electron acceptor and can oxidize NADH to NAD+, which is then reused to re-oxidize glucose.
Image: Karl-Erik Johansson (BVF, SLU) - Click on the image to enlarge it.
Introduction
All bacteria of importance in veterinary and human medicine are chemoorganoheterotrophic, which means that they get their energy from organic carbon compounds, which are also used as carbon source. The only exception is cyanobacteria, which are photolithoautotrophic, i.e. they get their energy from light and they use inorganic carbon in the form of CO2 (carbon dioxide) as carbon source.
Bacteria can thus extract energy through oxidation of carbohydrates (especially glucose) and these bacteria are said to be saccharolytic. Some other bacteria can instead use amino acids or lipids as an energy source, but this is less common and these bacteria are said to be asaccharolytic. Carbohydrates are also included in a variety of cellular processes in bacteria and bacterial carbohydrate metabolism is highly diversified. This is the reason why bacteria can grow and thrive in practically every environment on our planet.
Glycolysis
Glucose, which is a 6-carbon monosaccharide (hexose monosaccharide), is one of the most important carbohydrate molecules that bacteria can extract energy from. This is done by an oxidation process, called glycolysis, and in the glycolysis, glucose is broken down into pyruvate. Bacteria can also break down e.g. lactose and mannitol to pyruvate by glycolysis. Lactose is a disaccharide, which consists of two hexose monosaccharides (galactose and glucose). Mannitol is a hexose monosaccharide alcohol. If a bacterium has the right enzyme systems, then these carbohydrates can also be oxidized by glycolysis.
Under anaerobic conditions, pyruvate, can be converted to a variety of products, depending on the enzyme system of the microorganism in question. This process is called fermentation and by examining which carbohydrates a particular bacterium can ferment and which end products are formed, one can identify the bacterium in question.
Some bacteria can also convert the amino acid tryptophan to pyruvate if they have the enzyme typtophanase (can be detected by the indole test). Puruvat can then take various pathways in the metabolism, depending on whether O2 (oxygen) is available and depending on which enzyme system the bacterium has. If O2 is missing and if the bacterium is facultatively anaerobic, acid tolerant or anaerobic, puruvate can be further converted by fermentation.
Fermentation
Fermentation is a metabolic process that does not require O2 and can be utilized by many different bacteria. The fermentation in combination with the glycolysis results in the production of ATP, which is the most important form of energy for bacteria and other organisms. Examples of end products after the fermentation are: lactic acid (lactobacilli), ethanol (yeast), 2,3-butanediol (Klebsiella spp.), formic acid (Shigella spp.), butyric acid (Clostridium sp.) and mixed acids (E. coli m. al.). Glycolysis in combination with fermentation is not an efficient process for recovering energy because the carbohydrate is not completely oxidized to CO2 and H2O. Therefore, only 2 ATP molecules are formed for each carbohydrate molecule that is oxidized and facultative anaerobes always grow better in the presence of O2.
In the sections on the Krebs Cycle and on the Electron transport chain it is described how the chemical energy in pyruvate can be more efficiently extracted.
Updated: 2019-10-10.
Gram staining
Introduction
Gram staining is a so called differential staining techniques, since one can distinguish two major groups of bacteria by this method. These two groups are gram positive and gram negative bacteria, which are stained purple and pink to red, respectively.
Principle
Gram-positive bacteria have a thick cell wall (peptidoglycan), which consists of several layers and can be likened to a network. Gram negative bacteria have a much thinner cell wall and also an outer membrane. Crystal violet (CV+), which is the primary dye binds to the negatively charged groups on the bacteria and stain them purple. Then iodine (I-) will be used to form a large complex (CV-I) with CV and thereby bind the stain to the bacterium. When Gram-positive bacteria are treated with the decolourizing solution (ethanol-acetone), the bacteria will be dehydrated and the colour retained. When Gram-negative bacteria are treated with the decolourizing solution the outer membrane will be dissolved and the thin peptidoglycan exposed, so that the CV-I complex is washed out. Then a counterstaining with safranin or basic fuchsin is performed to stain gram negative bacteria pink or red.
Method
- Label the slide with the help of the diamond pen to mark which side of the slide that bacteria will be.
- Disperse some colony material in a drop of NaCl, air dry (in the incubator, if necessary).
- Fix the specimen by moving the slide, with the preparation side up, 6-8 times through the burner flame.
- Add chrystal violet, wait for 1 minute.
- Flood gently with Lugol's solution.
- Add Lugol's solution (which contains iodine), wait for 1 minute.
- Flood gently with acetone-ethanol solution.
- Flood gently with water.
- Add safranin, wait for 20 seconds.
- Flood gently with water.
- Remove the excess of fluid with a paper towel and allow to air dry until the specimen is completely dry.
Gram-positiva bacteria
Members of the phyla Bacillota and Actinomycetota (exception: genus Mycobacterium).
Gram-negativa bacteria
Members of the phyla Bacteroidota, Campylobacterota, Cyanobacteria, Fusobacteriota, Pseudomonadota (exception: some members of the order Rickettsiales), Spirochaetota and Thermodesulfobacteriota.
Updated: 2025-05-23.
Growth curve
Introduction
Bacteria multiply by binary fission (two identical daughter cells are formed upon the cell division). If the availability of nutrient is constant, and the physico-chemical properties of the culture medium does not change, they divide by the same speed all the time. This means that growth is exponential. The exponential growth phase (log phase) can not continue indefinitely, because the culture medium will become depleted of nutrients and the pH of the medium usually changes. When a bacterial culture is initiated in a liquid medium, it takes some time before the bacteria will start to grow, especially from clinical specimens or from hypothermic cultures and this phase of cultivation is known as the lag phase.
Generations time
The time required for the number of bacteria to double during cultivation is called generation time or doubling time and it can vary greatly for different bacteria. Escherichia coli, which is cultivated under optimum conditions, has a generation time of 20 min, whereas Mycobacterium avium subsp. paratuberculosis has a generation time of about 24 hours. This means in practice that if you spread these two bacteria on appropriate culture plates, you can easily see colonies of E. coli after one day, while colonies of M. avium subsp. paratuberculosis cannot be observed until after at least 3 months!
Growth curve
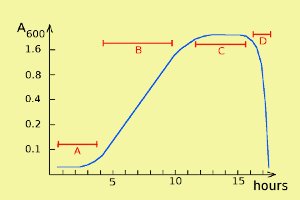 Bacterial growth can be described by a so-called growth curve with four different phases: lag phase, log phase, stationary phase and death phase (= decline phase) (labelled A, B, C and D, respectively, in the figure). Note that the scale of the y axis is logaritmic. During the stationary phase bacteria die at about the same rate as they are formed during cell divisions, and in the death phase, bacteria die faster than they regenerate.
Bacterial growth can be described by a so-called growth curve with four different phases: lag phase, log phase, stationary phase and death phase (= decline phase) (labelled A, B, C and D, respectively, in the figure). Note that the scale of the y axis is logaritmic. During the stationary phase bacteria die at about the same rate as they are formed during cell divisions, and in the death phase, bacteria die faster than they regenerate.
Construction of a growth curve
When a growth curve is constructed, you most often want to be able to follow the growth of bacteria in real time to know when to make e.g. addition of a substance, or when cultivation should be interrupted. The fastest and easiest method is to regularly collect samples from the culture and determine light scattering by spectrophotometry. With a spectrophotometer you normally determine absorbance at different wavelengths, but in a suspension (e.g. of bacteria) light scattering (OD = optical density) can also be measured. Light scattering is proportional to the number of bacteria per ml, and by means of a standard curve, one can determine the number of bacteria in absolute terms.
Updated: 2025-05-23.
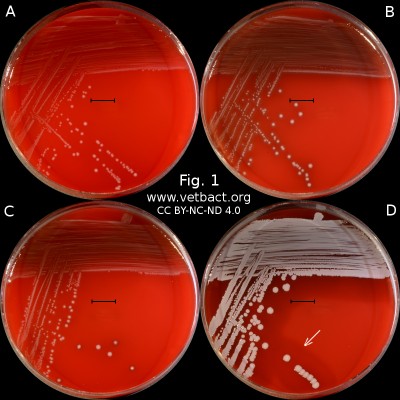
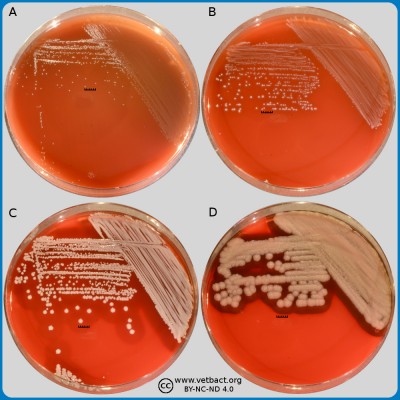
Hemolysis
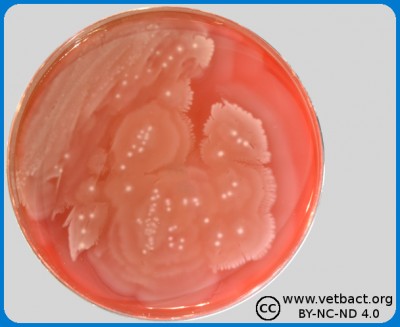
Fig. 1. The image shows colonies of some bacterial species that exhibit different hemolys patterns. The colonies have been lit from above during photography. However, the easiest way to observe haemolysis is with the illumination from below and by looking at the plate in the "right" angle. The following bacteria have been used to illustrate hemolysis:
A. Streptococcus uberis, causing no hemolysis. This is sometimes is called γ-hemolysis, which is a bit unfortunate.
B. Streptococcus agalactiae, which gives a clear (complete) β-hemolysis.
C. Streptococcus dysgalactiae (subspecies not defined), that gives incomplete greenish α-hemolysis.
D. Staphylococcus pseudintermedius, giving double hemolysis.
- Click on the image to enlarge it.
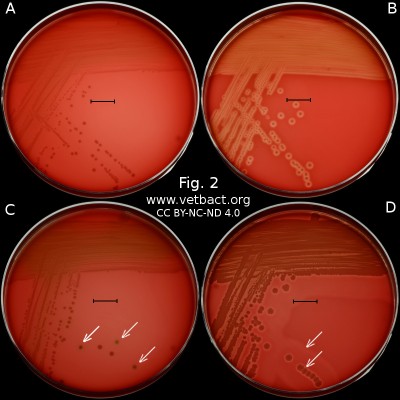
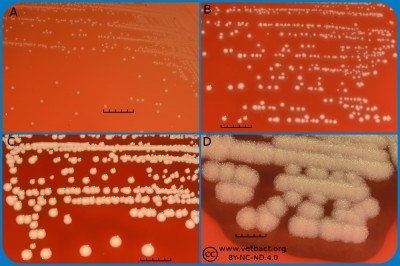
Fig. 2. This image shows the same agar plates as in Fig. 1, but the colonies have been lit from below during photography, because it is the easiest way to observe the haemolysis. - Click on the image to enlarge it.
Introduction
Hemolysis (Brittish spelling: haemolysis) means that the red blood cells (erythrocytes) burst apart (hemolyse) and release the cell contents (hemoglobin). Some bacteria produce so-called hemolysins, which give them hemolytic capacity. Most hemolysins are proteins (enzymes or porins), but there are also other types of hemolysins like rhamnolipids and biological detergents (biosurfactants).
Protein hemolysins
Hemolysins are membrane distupting exotoxins which can be divided into two groups: toxins with enzymatic activity, and channel-forming toxiner (= porins).
Enzymatically active hemolysins are often lipases such as α-toxin of Clostridium perfringens, which is a phospholipase. When lipase cleaves lipids in plasma membranes of the host animal cells, the membrane will become fragmented and the cell contents leak out.
Porins are composed of subunits, but are secreted by the bacterium in monomeric form. In the cell membranes of the host animal, the monomers aggregate to channel-forming polymers (heptamers), which makes it impossible for the ion gradient across the plasma membrane of the host cell to be maintained and the osmotic pressure in the cell will increase until it lyses.
Function
A function of hemolysins is that the bacteria can utilize hemolysis to release and utilize nutrients from the host animal cells. Iron e.g., is essential to many pathogenic bacteria, but is only present in very low concentrations outside the cells. If the bacteria have access to free hemoglobin, it can utilize the iron, which is bound to the heme groups of hemoglobin. Hemolysins do not act only on erythrocytes, but can also lyse other types of cells.
Identification of bacteria based on haemolysis
By cultivation on blood agar, bacteria can be differentiated based on their capacity to secrete hemolysins. The hemolysis will cause a clearing zone of the blood agar around the colonies. Bacteria can cause different types of hemolysis:
- α-hemolysis, which means an incomplete clearing (green haemolysis).
- β-hemolysis, which means a complete clearing.
- Double hemolysis of some staphylococci consisting of an inner β-hemolysis zone and an outer α-hemolysis zone (see also below).
- No hemolysis, which is sometimes referred to as γ-hemolysis, which may seem illogical.
Note that the α-hemolysin of staphylococci causes complete hemolysis, whereas their β-hemolysin causes incomplete hemolysis.
The capacity to produce hemolysins may vary between different strains of a particular bacterial species. Blood agar plates with bacteria having different hemolysis patterns are shown in Fig. 1 and 2. Note that in Fig. 1A and 2A hemolysis cannot be observerd, but note that there are other strains of S. uberis that give α-hemolysis . In Fig. 1B, one can discern a thin hemolysis zone and in Fig. 2B, the clear β-hemolysis is evident around all colonies. In Fig. 1C it is possible to discern hemolysis around some colonies, and in Fig. 2C, one can clearly see the green α-hemolysis around some colonies (white arrows). In Fig. 1D one can see the outer hemolysis zone (white arrow) and in Fig. 2D, it is possible to see both the clear inner β-hemolysis zone and outer turbid α-hemolysis zone (white arrows).
Note also that all the colonies of Streptococcus dysgalactiae do not give rise to α-hemolysis, although the strain used is pure with respect to species. However, it may be that different strains (or clones) of the same species exhibit different hemolysis patterns.
Updated: 2025-05-23.
Homogenization
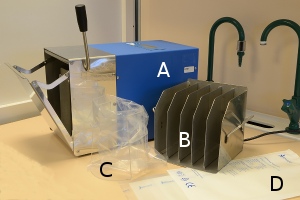 In order to cultivate bacteria from certain types of samples (including food and feed) it is important to disperse the sample in the dilution broth without damaging the bacteria. A Stomacher® is an apparatus used for homogenisation of samples in food microbiology. A large amount of viable microorganisms can be released from different kinds of food samples by this method. The apparatus does not have to be sterilized because the samples are contained in sterile plastic bags to which sterile medium is added.
In order to cultivate bacteria from certain types of samples (including food and feed) it is important to disperse the sample in the dilution broth without damaging the bacteria. A Stomacher® is an apparatus used for homogenisation of samples in food microbiology. A large amount of viable microorganisms can be released from different kinds of food samples by this method. The apparatus does not have to be sterilized because the samples are contained in sterile plastic bags to which sterile medium is added.
A. The homogeniser, which is loaded with a plastic bag containing the sample and culture fluid
B. Rack for the plastic bags during the incubation
C. Rack, which is used during the weighing of sample material and addition of culture medium
D. Sterile plastic bags in the homogenizer and for the incubation
Click on the image to enlarge it.
Updated: 2025-05-23.
Infection vs. inflammation
Introduction
Infection and inflammation are two concepts that are often linked, but they are not synonymous.
Infection
Infection means that tissues in the body have been colonized by microorganisms (bacteria, fungi, viruses or protozoa), which continue to multiply and cause tissue damage. If the microorganism in question does not cause damage, but belongs to the normal flora and is perhaps even useful for its host animal, one should instead speak about colonization.
Inflammation
Inflammation is the body's defense reaction to an attack of microorganisms or an injury (eg, stroke, heat, cold or allergy). Some diseases can also cause inflammation. The body's defense reactions are characterized by, among other things, the peripheral blood vessels being widened to become permeable to blood plasma and immune cells (white blood cells), which can then leave the bloodstream and seek out and disarm microorganisms or repair damage to the body's cells.
Infestation
In the case of ectoparasites, one sometimes sees the concept of infestation instead of infection and, particularly in American literature. However, this expression should be avoided as it may cause confusion.
Updated: 2019-03-04.
Iron
Why is iron essential?
Iron is essential for almost all forms of life, because it is a necessary component in for energy production, DNA replication, transportation of oxygen and protection against oxidative stress. Bacteria are not an exception in this respect and they have to be able to aquire iron in the animal hosts to multiply and cause disease.
How can the host animal utilize that iron is essential for bacteria?
One defense mechanism that higher organisms have against bacteria is to minimize the access to free iron. The main part of the iron in vertebrates is intracellular and bound to the iron binding protein ferritin or complexed to the porphyrin ring of heme, which is a cofactor to hemoglobin and myoglobin. Furthermore, in serum there is a iron transporting protein in serum, which is called transferrin, to which iron is bound extremely strong.
How can bacteria circumvent the animal host?
It wouldn't be possible for bacteria to survive within an animal host unless it has developed special mechanisms for extraction of the bound iron. Most pathogenic bacteria have mechanisms for uptake of iron with such high affinity for iron they can compete with the iron binding substanses of the host animal. These mechanisms can be based on siderophores, heme aquisitation or transferrin/lactoferrin receptors.
Borrelia burgdorferi, which causes Lyme disease, is an exception from the above rule since it utilizes manganese instead of iron. That is, enzymes which normally contain iron are modified and in B. burgdorferi they contain manganese. Members of the genus Lactobacillus have also been regarded as non-dependent of iron, but this has been questioned in recent studies.
Updated: 2017-11-10.
Koch's postulates
Introduction
Robert Koch was a German physician (1843-1910), who was interested in the relationship between microorganisms and disease. Koch formulated four criteria (Koch's postulates), which must be met to prove that a particular microorganism has caused a certain disease (viruses however, require other criteria).
Koch's postulates
- The microorganism must/should be detected in large numbers in all individuals who suffer from the disease, but it must/should not be present in healthy individuals
- The microorganism must/should be cultivated in pure culture from samples coming from the sick individual.
- The cultured microorganism must/should cause the sama disease in healthy individuals (laboratory animals).
- The microorganism must/should then be cultivated from or detected in the diseased laboratory animal.
In the original version of the postulates must was used, but with today's knowledge, one must use should as there are many exceptions. The first criterion had to be abandoned when it was discovered that there are asymptomatic carriers of certain microorganisms (subclinical infections). The second criterion must sometimes be abandoned because there are microorganisms that cannot be cultivated. The third criterion is not always valid because external factors can affect the results of an experimental infection.
Conclusion
If all criteria are met, a relation between microorganisms and disease has been proven, but if not all criteria are met, then it may still be a connection.
A number of new criteria to prove a relationship between microorganisms and disease was published in 1996 (Fredricks and Relman, Clin. Microbiol. Rev. 9:18-33). These criteria are based on detection of specific DNA sequences associated with disease.
Updated: 2017-09-27.
Krebs cycle
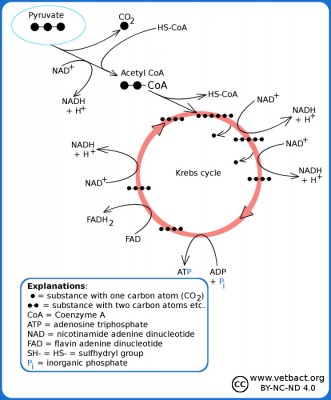
Simplified picure of the Krebs cycle where you can see how pyruvate is converted to acetyl-CoA, which reacts with oxaloacetate and forms citrate (hence the alternative name the citric acid cycle). From citrate, carbon atoms are split off in the form of CO2 and after a revolution in the Krebs cycle oxaloacetate remains, which can then take care of a new molecule of acetyl-CoA.
Image: Karl-Erik Johansson (BVF, SLU). - Click on the image to enlarge it.
Introduction
From one glucose molecule having 6 carbon atoms, two molecules of pyruvate having 3 carbon atoms each are formed during the glycolysis. Under aerobic conditions and if the current bacterium has aerobic metabolism, the pyruvate molecule is not fermented, but can instead be converted to acetyl Coenzyme A (acteyl-CoA), while one molecule of NAD + is reduced to NADH. The acetyl group in acetyl-CoA has two carbon atoms because carbon dioxide (CO2) has been cleaved off and can now enter the Krebs cycle (= citric acid cycle). The Krebs cycle is a prestep to the elektron transport chain (= the respiratory chain).
Krebs cycle
All enzymes required for the Krebs cycle exist in bacteria in the cytoplasm (= cytosol), as bacteria lack mitochondria. The acetyl group binds in the first step of the Krebs cycle to oxaloacetate, which has four carbon atoms, and then citrate, which has six carbon atoms, is formed. After one revolution in the Krebs cycle, 2 molecules of CO2 have been cleaved off, 3 molecules of NADH, 1 molecule of FADH2 and 1 molecule of ATP have been generated from each pyruvate molecule. What remains of the citrate molecule is again oxaloacetate, which can be re-acetylated and spin one more revolution in the cycle. In total, 8 molecules of NADH, 2 molecules of FADH2 and 2 molecules of ATP have been formed from 1 molecule of glucose. Actually, it is not ATP that is formed, but GTP. However, GTP is then converted in the cell to ATP by a reacton with ADP. NADH and FADH2 are utilized in the electron transport chain for ATP synthesis.
Other molecules than carbohydrates
Some bacteria have an enzyme (tryptophanase), which can form pyruvate from amino acids (tryptophan). Pyruvate can then be metabolized in the Krebs cycle. Other bacteria can break down cysteine into substances that can enter the Krebs cycle. In order for amino acids to be used in the Krebs cycle, they must first be deaminated, i.e. the amine group must be converted to ammonia (NH3) or rather to an ammonium ion (NH4+), which can be secreted. In addition, there are bacteria that can metabolize lipids to glycerol and fatty acids. Glycerol can be converted to pyruvate and further metabolised in the Krebs cycle and fatty acids can enter the Krebs cycle directly. Thus, there are many pathways into the Krebs cycle and bacteria with aerobic, facultative anaerobic or microaerophilic metabolism can, therefore, extract energy from many different types of substances.
Updated: 2019-10-10.
Lancefield grouping of streptococci
Streptococci are sometimes classified in so-called Lancefield groups after the American microbiologist Rebecca Lancefield (1895 - 1981), who developed a system for serological classification based on the carbohydrate composition of the cell wall. The different groups are called Lancefield groups A to V (except I and J). There is also a group NG (= non-groupable).
Updated: 2017-09-17.
Lipopolysaccharide (LPS)
Schematic image of a lipopolysaccharide (LPS). Variations in structure occur in different bacterial species. The core can be divided into an inner and an outer part. The inner portion consists of 1-3 KDO residues (keto deoxy octulosonate) and 2-3 heptose residues. The outer part consists of several different hexose residues (e.g. D-glucose, D-mannose and D-galactose).
Bild: Karl-Erik Johansson (BVF, SLU) - Click on the image to enlarge it.
Introduction
Lipopolysaccharides (LPS), which are also known as lipoglycans, are only present in the outer membrane of gram negative bacteria. LPS is a so-called endotoxin (se även toxin nedan) and the toxicity is associated to lipid A (see below).
Structure
LPS is an amphipathic molecule, i.e. it has both hydrophilic (water loving) and hydrophobic (water repellent = lipid loving) regions. The hydrophobic portion (hydrocarbon chains) anchors LPS in the outer lipid layer of the bacterial outer membrane and the hydrophilic region (charged groups) points outward against the bacterial environment. Chemically, LPS consists of a lipid moiety and a polysaccharide moiety. The lipid moiety comprises lipid A, which is a phosphorylated glucosamine disaccharide with several (4-6) linked hydrocarbon chains, which constitute the hydrophobic portion of the molecule. The carbohydrate moiety consists of a so-called core oligosackaride (core antigen or R antigen), which is directly bound to lipid A. The polysaccharide (O-polysaccharide, O-antigen or somatic antigen) is bound to the R antigen.
Specificity
Thanks to the diversity, found in LPS from various gram-negative bacteria, its antigenic properties can be used for typing and subtyping of bacteria. The antigenic properties will also vary in different parts of LPS:
- Lipid A is often family-specific. In for instance the family Enterobacteriaceae, there is almost no variation in the structure of lipid A.
- The R antigen is often specific to the genus, i.e. no significant variation occurs in this part within a single bacterial genus.
- The O-antigen is not even species specific, i.e. considerable variation occurs even within a species and this provides opportunities for subtyping by serological methods. Thus, one can distinguish between different strains of the same species, which makes it possible to use serology for epidemiological studies.
Effects on the host animal
Animals (including humans) are constantly exposed to small amounts of LPS in the blood circulation because of the turnover of intestinal gram negative bacteria and thus the innate immune system is contiuously stimulated. Exposure to large amounts of LPS, as during for instance sepsis, will cause release of cytokines, leading to fever and inflammation. At toxic concentrations of LPS, blood clots form in the capillary system, which in turn can lead to life-threatening conditions.
Updated: 2020-03-19.
Logarithms and dilutions
Introduction
Logarithms (log) are often used to simplify calculations with large numbers. When discussing the number of microorganisms in cultures from for instance food samples or when preparing and describing dilutions, logarithms are most often used to express the dilution factor.
Calculations
The logarithm (log) of a certain number (x) is the number (y) to which another number (b) must be raised, to give the number x. That is:
x = by
The number b is called the base in the logarithmic system you want to use and most common in science and engineering is that you use the number 10 as a base and then you talk about base-10, decimal or common logarithms, which are usually written log10 or lg10. For the sake of simplicity, only log or lg is often written or said, when referring to the base-10 logarithm. An example:
lg 1000 = 3 because 1000 = 10x10x10, which can also be written 103.
According to the logarithm rules, e.g. the following expressions apply for all types of logarithms:
- log (a x b) = log a + log b
- log (a / b) = log a - log b
- log ap = p log a
It follows from the second rule that log10 (1 / b) = 0-log10b = -log10b
I.e. logarithms can also be negative, e.g.:
log10 (1/1000) = -3, since log10 (1/1000) = 0-3 = -3
In other words:
...
103 = 1000
102 = 100
101 = 10
100 = 1 (can not really be explained mathematically, but has for practical reasons been defined as 1) )
10-1 = 0.1
10-2 = 0,01
10-3 = 0.001
...
Dilutions
In microbiology, one often wants to make dilution series when deciding how many microorganisms there are in a sample. The most common is to make a 10-step dilution series to achieve high accuracy at large dilutions. This means that the dilution factor is 1:10 in each step and that from one sample you take 1 part of sample solution to 9 parts of culture medium (or another suitable liquid) in a test tube, mix and transfer 1 part of mixture to 9 parts of culture medium in a new test tube and so on until a suitable degree of dilution has been reached. Then you will get a series of test tubes with dilution factors 1:10, 1: 100, 1: 1000, 1:10 000 and so on. These dilution factors can also be written as: 1/10, 1/100, 1/1000, 1/10 000 and so on, or 0.1, 0.01, 0.001, 0.0001. This can also be expressed with the help of logarithms, i.e. 10-1, 10-2, 10-3, 10-4 and so on.
Therefore, a 1:1000 dilution is sometimes casually referred to as a minus-three dilution.
Read more about how to use dilution series to determine bacterial amounts under Colony Forming Unit (CFU).
Other logarithms
Logarithms with bases other than 10 are used for practical reasons in different disciplines.
Binary logarithms (lb) with base 2 are used in e.g. computer science, bioinformatics and photography.
Natural logarithms (ln) with base e [a transcendental number (limit value), which is approximately equal to 2.71828 ...] are used in e.g. mathematics, physics and chemistry.
Updated: 2021-06-23.
Macromorphology
The figure shows blood agar plates with four different bacteria, which have been incubated during 24 h at 37°C. These bacteria (A, Erysipelothrix rhusipathiae; B, Streptococcus agalactiae; C, Staphylococcus aureus; D, Bacillus cereus) represent four different colony sizes. The length of the scale bar is equivalent to 5 mm. - Click on the image to enlarge it.
The appearance of a bacterial colony on agar plates depends on, among other things, growth medium, nutritional availability, incubation time and temperature. There are many different culture media used for typing and/or selective cultivation of bacteria. One single bacterial species could, thus, have different appearance depending on the medium in the agar plate. The appearance of a bacterial colony is usually judged on a blood agar plate.
A bacterial colony usually originate from a single bacterium, but once it becomes visible it consists of several miljons of bacterial cells. Such a collection of bacteria is usually referred to as a colony forming unit (CFU), because you cannot know for certain that it originates from one single bacterial cell. Bacterial colonies differ in appearance regarding size, colour and shape etc. The bacteria included in VetBact are classified according the size of the colonies they form:
- Pin-point sized colonies (<1 mm in diameter), A in the figures
- Small colonies (1-2 mm in diameter), B in the figures
- Medium sized colonies (2-3 mm in diameter), C in the figures
- Large colonies (>3 mm in diameter), D in the figures
Other macromorphological characteristics of bacterial colonies that can be used for identification/classification, are the following:
The entire bacterial colony
- Circular (round)
- Irregular
- Swarming
- Mucoid
The shape of the margin of the colony
- Entire
- Irregular (undulate or lobate)
- Filamentous
The structure of the colony
- Smooth
- Wrinkled
- Wavy
The surface of the colony
- Shiny or dull
- Moist or dry
The profile of the colony
- Flat and low
- Flat and high
- Convex
- Cupol shaped
- Peak shaped
Furthermore, the following characteristics can also be used for identification/classification: for instance colour, apacity, odour and possible hemolysis.
Updated: 2018-06-28.
Matrix-Assisted Laser Desorption/Ionization Time Of Flight Mass Spectrometry (MALDI-TOF MS)
 The instrument in the image belongs to the Department Animal Biosciences at the Swedish University of Agriculture Science (SLU). Marina Falk (018-672389) is responsible for the equipment and it is possible to get isolates analyzed. - Click on the image to enlarge it.
The instrument in the image belongs to the Department Animal Biosciences at the Swedish University of Agriculture Science (SLU). Marina Falk (018-672389) is responsible for the equipment and it is possible to get isolates analyzed. - Click on the image to enlarge it.
Schematic description of the principle of MALDI-TOF MS. - Click on the image to enlarge it.
The principle of MALDI-TOF
Mass spectrometry based on the MALDI-TOF (or Maldi only) means that the bacterial isolate to be analyzed, is adsorbed to some type of carrier material (matrix). The isolate is then irradiated with laser UV light, so that the molecules in the bacteria are broken into positively charged fragments (ionization), which are thrown towards a detector. The time it takes for the fragment to reach the detector (time of flight) is measured. The time is dependent on fragment size and charge. Also very large molecules (proteins and nucleic acids) can be fragmented and ionized in this way. Large molecules give rise to many fragments and a characteristic mass spectrum, which can be used for identification.
The use of MALDI-TOF MS for identification of bacteria
One can perform these analyzes directly on bacterial colonies and the resulting mass spectrum is then compared with stored mass spectra of known bacterial species. Thus, you will get an analytical response within a minute and the method is considered to be very reliable. The more mass spectra of known bacteria you have to compare with, the safer the method will be.
MALDI-TOF MS is already used in some laboratories for veterinary bacteriology and many researchers believe that this technique will be tomorrow's routine method for identification of bacteria. The instrument is still very expensive, but material costs are low.
Score value
The software for the Maldi system contains a database with information about which fragments one can possibly get from a particular bacterium. This information can be used by the system to compare with unknown isolates and calculate a score value for each isolate. The score value is a measure of the probability that a particular isolate will represent a particular bacterial species/strain in the reference library. A score value between 0.000 and 1.699 means that identification is not possible since the peak of the unknown isolate does not represent any bacterium in the reference library. A score value between 1.700 and 1.999 are reliable only to the genus level, but a low confidence of identification at species level compared to reference peak list. . A score value between 2.000 and 3.000 are considered accurate of both genus- and species-level.
Updated: 2024-12-09.
Microaerobic bacteria
Microaerobic incubation by commercial gas-generating envelopes in sealed jars. - Click on the image to enlarge it.
Background
Microaerophilic bacteria require a reduced oxygen concentration for growth. They have optimal growth in 5-10% oxygen and some species are stimulted by 1-10% carbon dioxide. Bacteria that thrive in the precence of carbon dioxide are called capnophilic bacteria (or capnophiles).
The normal atmosphere consists of about 21% oxygen. This concentration is toxic for microaerophilic bacteria, which might lack efficient enzymes (catalase or peroxidase and superoxide dismutase) to detoxify hydrogen peroxide and superoxide radicals, which are produced when oxygen is dissolved in water (see anaerobic bacteria).
The growth of some species is enhanced by the presence of hydrogen, but the reason for this has not been clarified.
Microaerobic incubation
The cultivation of microaerophilic bacteria requires special laboratory technique. Several methods are used to generate microaerobic atmosphere. Microaerophilic bacteria could be culitivated in sealed jars or plastic bags with commercial gas-generating envelopes. Special incubators or sealed jars with valves for evacuation of the normal atmosphere and refilling of the optimal gas mixture can also be used.
Example Microaerophilic bacteria
Campylobacter spp., Borrlia spp. and Helicobacter spp.
Updated: 2020-02-14.
Motility
Introduction
Many bacteria are motile and they can migrate by utilizing mechanisms based on different principles. Motility mechanisms have been developed in bacteria in order for them to be to be attracted or repelled by certain stimuli. Motility is a feature, which is used for characterization and identification of bacteria and methods have, therefore, been developed to detect motility. Bacterial locomotion must be distinguished from so-called Brownian molecular motion, which is random and depends on the thermal motion of water molecules, which can push the bacteria so that it is perceived as if the bacteria move under the microscope.
Swimming movement by means of flagella
Video clip displaying the motility of Brachyspira murdochii (Spirochaetota).
Bacteria can have flagella, which may be one or several in number, and which act as propellers and allow the bacteria to move in a predetermined direction. Very simplified, you can say that the flagellum consists of a moving filament, which is linked to a molecular engine (basal body) in the bacterial envelope via a hook. The flagellum consists of many subunits of the protein flagellin, which form a hollow and flexible cylinder. The molecular engine, which is built up by protein subunits, is driven by the proton gradient (or a Na+ ion gradient) over the cell membrane and can get the filaments to rotate clockwise or counterclockwise. This rotation causes the bacteria to swim in a certain direction and to tumble, respectively. The direction is dependent on outer stimuli and during tumbling, the bacteria can change direction of the movement.
Bacteria within the phylum Spirochaetota have so-called periplasmic flagella (endoflagella or axial filaments), which are localized in the periplasmic space between the cell membrane and the outer membrane. This arrangement result in a screw like motility (or flat-wave motility), which makes it possible for these bacteria to move in highly viscous material (like mucus).
Twitching motility by means of type IV pili (fimbria)
Bacteria which have so-called type IV pili can move by using the external ends of the pili, which have hooks, and can adhere to a solid substrates like the surface which the bacteria colonize or to other bacteria. When the pilus contracts, the bacteria are pulled forward. Movement produced by type IV pili is typically jerky, and thus it is simply called twitching motility. Pseudomonas aeruginosa has this twitching motility.
Gliding motility on surfaces
Some bacteria can glide on wet surfaces, but the molecular mechanisms are incompletely understood. Examples of bacteria, which have this ability are: members of the phylum Cyanobacteria and the genera Flavobacteria and Mycoplasma.
Actin-based motility
Some pathogenic bacteria can move inside the host cell by using its cytoskeleton. The cytoskeleton is normally used to move organelles inside the eukaryotic cell. By stimulating actin polymerization at one of its poles, these bacteria can form a kind of tail, which pushes them through the cytoplasm of the host cell. Examples of bacteria that can use this mechanism are species within the genera Burkholderia, Listeria, Rickettsia and Shigella.
Motility assay
Motility tests can be performed in test tubes containing a semi-solid medium, i.e a medium with a low concentration of agar. The medium is inoculated with a plastic loop, which is inserted into the agar and pulled up again. If the bacterium is motile, the medium in the whole tube will turn turbid, but if it is non-motile turbidity will only be obtained where the plastic loop was inserted. The picture shows a negative control (A), Staphylococcus aureus subsp. aureus (B) which is non-motile and Escherichia coli (C), which is motile.
Updated: 2025-07-01.
NAD and FAD
The picture shows the structure changes of FAD during oxidation/reduction. Note the two extra hydrogen atoms in the reduced form of FAD. The chemical structure of AMP and ribose are shown in the picture under ATP in the term list. - Click on the image to enlarge it.
The picture shows the structure changes of NAD during oxidation/reduction. Note the extra hydrogen atom in the reduced form of NAD. The chemical structure of ADP and ribose are shown in the picture under ATP in the term list. - Click on the image to enlarge it.
Nicotinamide adenine dinucleotide (NAD)
NAD is a dinucleotide consisting of an adenine moiety and a nicotinamide moiety which are joined by two phosphate groups between the ribose moieties of the respective nucleotide. NAD is present in an oxidized form (NAD+) and a reduced form (NADH) in all living cells and this is essential for all living organisms.
Functions
NAD participates in so-called redox processes in the cell, i.e. chemical reactions where one molecule is reduced and another molecule is oxidized. NAD+ is thus the oxidized form of NAD, which can oxidize other molecules and then itself be reduced to NADH by cathing electrons (e-). NADH can in turn reduce other molecules by emitting electrons. The most important function of NAD are these electron transferring reactions. NAD also has other functions, e.g. as substrates for enzymes, which catalyze modification of macromolecules ("post translational modification") by cleavage or binding of chemical groups to e.g. proteins.
Other
The so-called V-factor, which is needed for certain bacteria to grow in artificial media, is in fact nicotinamide adenine dinucleotide (NAD) or nicotinamide adenine dinucleotide phosphate (NADP, see below).
Nicotinamide adenine dinucleotide phosphate (NADP)
NADP has the same chemical structure as NAD and an additional phosphate group in the 2' position of the ribose moiety belonging to adenosine. NADP is used by the cell in anabolic (biosynthetic) processes.
Flavin-adenine dinucleotide (FAD)
FAD consists of two major components AMP and flavin mononucleotide (FMN), which are linked to each other by a so-called phosphodiester bond. The nucleotides in nucleic acids (DNA and RNA) are also linked to each other by such bonds. FMN is a derivative of riboflavin (= vitamin B2). Thanks to the flavin group, FAD can act as a cofactor for enzymes that catalyze certain complicated redox processes, e.g. in the Krebs cycle.
Updated: 2019-10-31.
Nomenclature of bacteria
Introduction
Nomenclature of bacteria refers to naming and bacteria and other organisms are named according to the binomial system, which was introduced by Carl Linnaeus (1674-1748). This means that a bacterium has a species name, which is composed of a genus name that tells you to which genus it belongs and a species epithet which, together with the genus name, is unique to the bacterium. An example of this is Moraxella bovis, where the genus name indicates that the bacterium belongs to the genus Moraxella and the species epithet indicates that the bacterium has been isolated from cattle. The genus name and the species epithet form together the scientific name of the species, which is always written in italics. Bacterial names are international and Latin or latinized Greek are used to form the name. If misunderstandings cannot occur, you can abbreviate the genus name after it has been written for the first time in a text, e.g. M. bovis. However, note that there are also bacteria called Mycoplasmopsis bovis and Mycobacterium bovis.
There are strict international rules for how bacteria should be named and these rules are published in a book named: "International Code of Nomenclature of Prokaryotes". In order to get a proposed name accepted, a scientific paper on the proposed species must be published and approved by an international taxonomy committee.
Trivial name
Trivial names are often used as a simplified way of naming a bacterial genus. A trivial name should neiter be written with capital first letter nor in italic. Examples of trivial names are: lactobacilli, mycobacteria, salmonella, staphylococci and streptococci. The scientific names for these groups are: genus Lactobacillus (or Lactobacillus spp.), genus Mycobacterium (or Mycobacterium spp.), genus Salmonella (or Salmonella spp.), genus Staphylococcus (or Staphylococcus spp.), genus Streptococcus (or Streptococcus spp.), respectively.
If you refer to a specific bacterial species, a trivial name refering to a complete genus should never be used.
Subspecies, biovars and serovars
Sometimes there is a need to divide bacterial species into subspecies, because they are too closely related to be regarded as different species, but too distantly related to be regarded as the same species. In this case a subspecies is introduced by adding a subspecies epithet and write subspecies (subsp. or ssp.) in front of it. An example of this is Streptococcus equi subsp. equi. When you divide a species into several subspecies, the original species always gets the same subspecies epithet as the species epithet.
There is often a need to divide species and subspecies in different biovars (biological variants) or different strains, but this is not strictly regulated, which means that researchers themselves can name their strains or biovars. One type of biovar is serovar (serological variant), by which various surface antigens can be identified with specific antibodies. Contact tracing and epidemiology is based on identification of different variants of the same bacterial species.
Serovar vs. serotype
Serovar and serotype are synonyms and thus, interchangeable terms, but according to the Rules of the Bacteriological Code (1990 Revision), serovar is the preferred term. Serogroup is a group of bacteria containing a common antigen. A serogroup may contain several serotypes. Serogroup is not an official designation, but has been used to classify bacteria belonging to the genera Leptospira, Salmonella, Shigella and Streptococcus.
Salmonella nomenclature
A bacterial subspecies that occurs in several thousand different serovars is Salmonella enterica subsp. enterica. A common serovar is Dublin and if you you want to write the complete and correct name of the bacterium, it becomes Salmonella enterica subsp. enterica serovar Dublin. Please note that the name of the serovar is capitalized, but not italicized. If the name appears in several places in the text, you can write S. enterica subsp. enterica serovar Dublin. However, because even this abbreviated writing is rather lengthy, it has been agreed that it is acceptable to simply write Salmonella Dublin, except on the first occurrance in a text, where the name must be given in full.
You can read more about naming of salmonellas on VetBact at Salmonella spp. and Salmonella enterica.
Updated: 2023-09-27.
Parameter
Parameter is a term, which is usually used to identify the variable and measurable characteristics, which define a system. The system may e.g. consist of a bacterial culture and examples of parameters are: number of added bacteria, growth temperature, incubation time, salt concentration, glucose concentration, CO2 concentration, etc.Testing parameter in food microbiology refers to standardized routine methods for the determination of e.g.: number of slow-growing bacteria, intestinal enterococci, culturable microorganisms etc.
Updated: 2013-02-26.
Pathogenicity
Introduction
The ability of a microorganism to cause any kind of damage (i.e. disease) to the host animal is called pathogenicity and microorganisms that have this ability are called pathogenes (or pathogenic organisms). Pathogenic is an "all-or-none-characteristic", i.e. a microorganism is either pathogenic or non-pathogenic in a given host animal. Virulence is not synonymous with pathogenicity, but describes the degree of damage that the pathogen has caused. A highly virulent bacterium is very contagious and/or gives severe symptoms.Obligate or opportunistic pathogens
A microorganism may be an obligate pathogen or an opportunistic pathogen. An obligate pathogen can be found in the host animal only in connection with disease. Microorganisms, which are found in healthy host animals, but which may cause a disease in certain circumstances are known as opportunistic pathogens. Such circumstances may be an immunocompromised host, other infection, tissue damage, etc.Characteristics that make a bacterium pathogenic
- Production of toxin.
- Production of adhesin.
- Production of capsule.
Updated: 2015-03-03.
Peptidogycan
The peptidoglycan in the cell wall of the bacteria is made up of repeating units consisting of two amino saccharides, N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). NAM has an oligopeptide consisting of 4-5 amino acids (aa) linked to it. The oligopeptide is also bound to another oligopeptide via an amide bond and thus all peptidoglycan chains are linked to a single large giant molecule.
Image: Karl-Erik Johansson (BVF, SLU). - Click on the image to enlarge it.
Introduction
Most gram-positive and gram-negative bacteria have a peptidoglycan in the cell envelope, which is 20-80 and 7-8 nm thick, respectively. The peptidoglycan is also called murein and is a major component of most bacteria's cell wall. The peptidoglycan is a polymer that forms a sparse mesh-like structure, which constitutes a single large three-dimensional giant molecule that stabilizes the cell envelope of the bacteria.
Composition
The peptidoglycan is made up of two alternating carbohydrate molecules containing amino groups: N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Each NAM molecule has an oligopeptide consisting of 4-5 amino acids linked to it. The oligopeptide from one peptidogycan chain is cross-linked to another oligopeptide from another peptidoglycan chain (see figure) via a so-called amide group (-CO-NH-). Amino acids (except glycine) can exist in two different isomeric forms, L-form and D-form, which are mirror images of each other. Living organisms usually contain only the L-form of amino acids, but the oligopeptide in the cell walls of bacteria consists of both the L- and D-form of the constituent amino acids as well as an amino acid (meso-diaminopimelic acid), which is not a part of ordinary proteins. Different species of bacteria may have slightly different composition of the oligopeptide.
Function
In addition to being a mechanical protection and a protection against osmotic lysis, the cell wall has an important function in cell division. The bacterium extends longitudinally before cell division and then the cell wal has to grow by synthesizing more of the peptidoglycan. When the bacterial cell then divides, the peptidoglycan must be reshaped to seal the two daughter cells.
Updated: 2020-04-02.
Phage therapy
Honey bees in a hive in Västeråker, Sweden. However, this hive does not suffer from American foulbrood.
Photo: Karl-Erik Johansson (BVF, SLU).
Lytic bacteriophages (phages) can be used to kill bacteria and, in principle, also to treat bacterial infectious diseases. Because more and more bacteria are becoming resistant to antibiotics that are used for treatment, the interest of using phage therapy has increased. However, there are both advantages and problems in using phage therapy for treatment. One advantage is that phages are extremely specific and the do not affect any other bacteria than the host species and of course not eukaryotic cells. This means the the microbial gut flora of the host animal is not disturbed by phage treatment. The high specificity is, however, also a drawback, since a phage is not only specific for a certain bacterial species, but also to only one single strain av that species. This means that a complete cocktail of phages containing types that are specific to all pathogenic strains of the actual species, are need for a successful treatment. Phage therapy is still in the experimental stage, but many researchers believe that it has great potential in both human and veterinary medicine, because results of some successful experiments have been published. For instance, American foulbrood in honey bee larvae, which is caused by Paenibacillus larvae, has been successfully treated by phage therapy.
Updated: 2018-05-30.
Phylogeny of bacteria
See Taxonomy and Phylogeny of bacteria.
Updated: 2023-04-12.
Plasmid
Plasmids constitute secondary genetic material in bacteria, which can be used if the need arises (see also Genome above). Plasmids can be present in one or several copies in a cell, as they replicate independently of the chromosome. A bacterium may carry multiple different plasmids.Function
Plasmids may contain genetic information, which is necessary for:- Antibiotic resistance.
- Synthesis of antibiotics such as streptomycin in Streptomyces spp.
- Synthesis of bacteriocins that are toxic to the strains of the same bacterial species, which do not contain the same plasmid. Example: colicines of E. coli.
- Conjugation [e.g. F (fertility) plasmid in E. coli].
- Synthesis of enzymes that degrade organic substances.
- Virulence genes (see Virulence factors), such as those of the genera Shigella, Salmonella and Yersinia. Virulence genes can also be found in so-called prophages, which is another type of secondary genetic material.
Practical use
Plasmids have been of great use in molecular biology including cloning of genes. Genetically modified plasmids, which contain only a few genes and a suitable cloning site, are then used.Updated: 2013-03-06.
Pure culture
Re-culture of a bacteria in a mixed culture of at least four different bacterial species - Click on the image to enlarge it.
Introduction
A clinical sample seldom contains bacteria in pure culture, they are usually found in a mixed flora. Thus, the pathogenic bacterial species is often isolated in combination with the normal bacterial flora during a bacterial infectious disease. To identify key bacteria in clinical samples, you have to have them in pure culture (see figure). A pure culture thus contains only one species of bacteria, and this can be accomplished by streaking (see below) onto agar plates.
Updated: 2025-05-13.
Pyogenic
A pyogenic (= suppurative or purulent) bacterium is a pus-forming bacterium. If mucus is also generated, the bacterium is called mucopurulent. Examples of pyogenic bacteria are Burkholderia mallei, Klebsiella pneumoniae, Staphylococcus aureus, Sta. epidermidis, Streptococcus pyogenes och Str. pneumoniae.Pyogenic bacteria may cause pyemia (see sepsis below).
Updated: 2013-03-06.
Quorum sensing and biofilm
Introduction
Quorum sensing is a system for signaling and response in a population of organisms (e.g. bacteria), regulated by the population size. In other words, the gene expression (= protein synthesis) of certain proteins can be regulated in response to changes in population density. Quorum comes from the Latin and means "of them", which refers to the minimum number present, as required for taking a decision.
Why do bacteria communicate with each other?
If very few bacteria are present in a particular area, it is a waste of energy to produce and secrete e.g. certain enzymes and, therefore, gene expression is regulated by quorum sensing. Formation of biofilm is also regulated by quorum sensing. Synthesis of the constituents of a biofilm is initiated when the bacterial population density has reached a certain level.
How do bacteria communicate with each other?
Bacteria use quorum sensing if necessary, to regulate gene expression in relation to the population size. The bacterium releases chemical messengers in the form of complex organic molecules (polypeptides in gram positive and N-acyl homoserine lactones in gram-negative bacteria) to achieve this. The concentration of signaling molecules is not high enough for the bacteria to communicate with each other in a population of planktonic bacteria (see biofilm). However, in a biofilm it is, and therefore they can communicate.
Links
- A website for Quorum Sensing that is recommended: "The quorum sensing site".
- Videoklipp at YouTube that is recommended: "Quorum sensing, bacteria talks".
Updated: 2021-01-21.
Salmonella antigens
Salmonella spp., have, like other members of the Enterobacterioceae family, three main types of antigens that can be used in serological diagnostics and for identification. These antigens are especially useful for identifying bacteria within the following taxa: Escherichia coli, Klebsiella spp., Salmonella spp. and Shigella spp. For epidemiological studies, Salmonella enterica subsp. enterica is divided into different serovars (= serovarians) depending on which set of antigens the strain in question is possessing. The word serotype is also used synonymously with serovar. The following three main types of antigens have been defined:
- Somatic antigens (= O antigens or cell wall antigens). These antigens consist of different oligosaccharides in the heat-stable part of the cell wall and are also called lipopolysaccharide (= LPS). There are more than 60 different O antigens in Salmonella spp.
- Surface antigen (= K antigens). These antigens are heat sensitive and consist of the capsular polysaccharides, which thus surround the cell wall and may cover O antigens. The Vi antigen is present in a few serovars.
- Flagell antigen (= H antigens). These antigens, which consist of proteins and are heat sensitive, are only found in flagella of motile members of the Enterobacteriaceae family. Motile Salmonella types produce two types of H antigen (termed phase 1 or phase 2 antigen.
The Kauffman-White's classification scheme is based on O and H antigens of Salmonella spp. More than 2500 serovars (serotypes) based on O and H antigens have been identified in Salmonella spp. For epidemiological studies, Salmonella isolates are first tested with a polyvalent O antiserum targeting all different serotypes. If the isolate is positive, it is further tested with monovalent O antisera targeting specific serotypes. If the isolate is negative with the polyvalent antiserum, it must then be heat treated to destroy any K antigen which can mask O antigens. Thereafter the test is repeated with the polyvalent antiserum and if the isolate is now postive, it has to be tested with the monovalent antisera as well. Similarly, the strain in question is also tested for different types of H antigens. The O antigen determines which serogroup a certain Salmonella isolate represents and in combination with the H antigen, the serovar (= serotype) can be defined.
Example: A salmonella type called Salmonella enterica subsp. enterica Serovar Dublin has the antigen set: 1.9.12:g,p:no phase 2, which means that it has:
- O antigens: 1, 9 and 12
- H antigens of type Phase 1: g and p
- H-antigens of type Phase 2: none
The antibodies used in the test are linked to very small latex beads, so that the latex spheres should form visible aggregates with the bacteria and this is what is called agglutination. A bacterium, which is positive in agglutination with polyvalent antisera against both the O and H antigen, is likely to represent a Salmonella isolate, but in order to do epidemiological studies (trace the infection) one has to determine which serovar the isolate belongs to.
Reference: Read more about the salmonella nomenclature on the bacterial pages of Salmonella spp. and Salmonella enterica on VetBact and about different serovars in the Term List of VetBact.
Updated: 2023-02-22.
Salmonella serovars
Introduction
The genus Salmonella is divided into two (or three) species: Salmonella bongori, Salmonella enterica (and possibly Salmonella subterranea), see also Salmonella spp. Salmonella enterica is divided into six subspecies, where Salmonella enterica subsp. enterica is of highest interest in human and veterinary medicine. S. enterica subsp. enterica is in turn divided into several hundred different serovars, based on their antigenic composition. The abbreviation S. for Salmonella may be used if there are no ambiguities in the text. The different serovars then get very long names, so there is a convention to abbreviate them in a certain way, where Salmonella (or S.) should be printed and in an italic fontit. The name of the serovar must be written in the regular font and in capital first letter. The first time the long scientific name is used in a text, however, it must be printed out in full. The table below lists some common serovars with full and abbreviated names and their most common host animals.
| Scientific name | Abbreviated name* | Animal species |
| S. enterica subsp. enterica Serovar Anatum | Salmonella Anatum | Cattle |
| S. enterica subsp. enterica Serovar Cholerasuis | Salmonella Cholerasuis | Pig, wild boar |
| S. enterica subsp. enterica Serovar Derby | Salmonella Derby | pig |
| S. enterica subsp. enterica Serovar Diarizonae | Salmonella Diarizonae | Sheep |
| S. enterica subsp. enterica Serovar Dublin | Salmonella Dublin | Cattle |
| S. enterica subsp. enterica Serovar Enteritidis | Salmonella Enteritidis | Poultry, hedgehog, human |
| S. enterica subsp. enterica Serovar Gallinarum | Salmonella Gallinarum | Poultry |
| S. enterica subsp. enterica Serovar Hadar | Salmonella Hadar | Poultry |
| S. enterica subsp. enterica Serovar Kentucky | Salmonella Kentucky | Poultry |
| S. enterica subsp. enterica Serovar Livingstone | Salmonella Livingstone | Poultry |
| S. enterica subsp. enterica Serovar Newport | Salmonella Newport | Poultry, pig, cattle, water birds |
| S. enterica subsp. enterica Serovar Paratyphi | Salmonella Paratyphi | Human |
| S. enterica subsp. enterica Serovar Pullorum | Salmonella Pullorum | Poultry |
| S. enterica subsp. enterica Serovar Senftenberg | Salmonella Senftenberg | Turkey |
| S. enterica subsp. enterica Serovar Typhi | Salmonella Typhi | Human |
| S. enterica subsp. enterica Serovar Typhimurium | Salmonella Typhimurium | Pig, poultry, cat, cattle, hedgehog, human |
------------------------------------------------------------------------
*You may also abbreviate Salmonella to S. here, if it can't be misunderstood, although it doesn't look so nice in text.
Please note that the most common serovar on a certain animal species and in a certain geographic area is not necessarily the most common serovar on the same animal species, but in a different geographic area.
Updated: 2023-04-05.
Secretion systems, bacterial
Introduction
A remarkable evolution of secretion systems to transport substances across the cell envelope (=cytoplasmic membrane, cell wall and possibly outer membrane) has occured in bacteria. Bacterial secretion systems are "nanomachines" in the form of protein complexes located to the cell membranes. In pathogenic bacteria, their function is to secrete different substances, so-called effector molecules, which are predominantly proteins, which allow the bacterium to colonize or invade the host cell. The effector molecules can also manipulate the host cell so that it is less likely to survive the bacterial infection, because they constitute toxins, adhesins, or degradation enzymes, etc. The secretion systems are, thus, very important virulence factors and they can be divided into different types depending on their composition, structure, mechanism and evolutionary relationship. Gram-negative and gram-positive bacteria can have (at least) eight and (at least) four different secretion systems, respectively, and there are two fundamentally different mechanisms (one-step and two-step mechanism).
Mechanisms
Through the one-step mechanism, the effector molecules are transported directly across the bacterial cell membrane(s) into the host cell and this can occur in both gram-negative and gram-positive bacteria. Through the two-step mechanism, the effector molecules are first transported through the plasma membrane to the periplasmic space of the bacterium and then by means of other protein complexes through the outer membrane and into the host cell. This can only happen in gram-negative bacteria.
Secretion across the plasma membrane
Secretion across the plasma membrane occurs in both gram-positive and gram-negative bacteria and there are three main systems for this type of transport:
- Sec (general secretion or GSP)
- SRP (signal recognition particle)
- Tat (twin arginine translocation)
Secretion across the outer membrane
Of course, secretion across the outer membrane only needs to occur in gram-negative bacteria and through either the one-step mechanism or the two-step mechanism.
Different principle types of secretion
The secretion systems are usually divided into the following seven main types, but this division is not complete and sometimes not entirely clear. Then there are also subtypes of these main types:
- Type I secretion system (T1SS or TOSS)
A simple one-step mechanism that requires only three different proteins. For instance Escherichia coli and Pseudomonas fluorescens possess this system. - Type II secretion system (T2SS)
A mechanism that only transports proteins over the outer membrane and generally transports the protein first by means of Sec across the cytoplasmic membrane. - Type III secretion system (T3SS or TTSS)
This type is the most complicated of all secretion systems described so far. It is a one-step mechanism utilizing a cannula-like protrusion (appendix) which can make holes in the host's cell membrane so that the secreted protein can be injected. This secretion system is also called injectosome because of its similarity to a syringe. Up to 25 different proteins are required to build up the injectosome and it has the same evolutionary origin as bacterial flagellas. Certain species within the following gram-negative bacterial genera possess T3SS, for instance Pseudomonas, Salmonella, Shigella, Vibrio and Yersinia. - Type IV secretion system (T4SS or TFSS)
Can translocate proteins through both a one-step mechanism and a two-step mechanism. Bordetella pertussis, Helicobacter pylori and Legionella pneumophila, for instance, possess T4SS. This system can also translocate DNA. - Type V secretion system (T5SS)
This secretion system can be divided into three main categories. - Type VI secretion system (T6SS)
This secretion system consists of a "phage-tail-nail-like injectosome reminiscent of T3SS. T6SS has structural and functional homology with the bacteriophage T4 and is required for the following bacteria to be virulent: Burkholderia mallei, Edwarsiella tarda, Francisella tularensis, Pseudomonas aeruginosa and Vibrio cholerae. - Type VII secretion system (T7SS)
This type of secretion system consists of a family of similar systems, which are specialized for bacteria that have specific lipids in their cell wall (eg, mycobacteria). Corynebacterium diphtheria and Nocardia sp. have been found to possess T7SS.
References
You can find nice pictures of the different bacterial secretion systems at the website SlideShare (www.slideshare.net) if you search for "bacterial secretion system".
Updated: 2020-02-27.
Sepsis
Sepsis (blood poisoning) is a potentially deadly medical condition with a whole-body inflammatory state [systemic inflammatory response syndrome (SIRS)], which is usually caused by a bacterial infection.Septicemia (septicaemia) is a related medical term referring to the presence of pathogenic organisms that multiply in the bloodstream, leading to sepsis.
Bacteremia (bacteraemia) is transiently presence of bacteria in the blood and this term is not equivalent to sepsis.
Pyemia (pyaemia) is a form of septicemia, which results in widespread abscesses of a metastatic nature. Pyemia is usually caused by the presence of pyogenic (pus-forming) bacteria in the blood.
Updated: 2012-12-11.
Siderophore
Introduktion
The word siderophore originates from Greek and means iron carrier. Siderophores are low molecular weight substances that have very high affinity for iron (Fe3+) and they bind iron by so-called chelation. Iron is essential for almost all forms of life, because it is a necessary component in the respiratory chain and for DNA synthesis. Many bacteria can secrete siderophores, which allows them to absorb iron even in an environment where the concentration of free iron is very low. The bacterium can then take up the iron, since it has receptors for siderophores in its surface. In mammals, iron is tightly bound to various proteins (e.g. ferritin, lactoferrin, hemoglobin and transferrin) and therefore, pathogenic bacteria bind iron in the form of soluble complexes with siderophores. Siderophores are considered as virulence factors and about 300 different siderophores from various bacteria had been described by year 2015.
Examples of siderophores
| Bacterium | Siderophore |
| Bacillus anthracis | Bacillibactin |
| Bacillus subtilis | Bacillibactin |
| Burkholderia pseudomallei | Malleobactin |
| Escherichia coli and some other enteric bacteria ... | Enterobactin |
| ... e.g. Salmonella spp. | Enterobactin |
| Some species within the genus Mycobacterium (e.g. M. tuberculosis) | Mykobactin |
| Pseudomonas aeruginosa | Pyochelin och pyoverdine |
| Yersinia enterocolitica | Yersiniabactin |
| Y. pestis | Yersiniabactin |
| Y. pseudotuberculosis | Yersiniabactin |
See also individual bacterial pages for more examples.
Updated: 2017-11-10.
Simple stain techniques
Simple stain methods are methods that utilize the fact that the negatively charged cytoplasm of bacteria attracts positively charged dyes like crystal violet and methylene blue. This can be used for rapid staining of specimen on microscopic slides after fixation with ethanol.By simple stain techniques, it is possible to determine whether a sample contains rods or cocci, but not if they are Gram-positive or Gram-negative. To determine whether the bacteria are Gram-positive or Gram-negative, one can use Gram staining, which is a differential stain method.
Updated: 2013-02-24.
Spores
Introduction
Spores in bacteria is not the same as spores of fungi. Bacteria do not use spores to reproduce, but to survive adverse conditions (lack of nutrients, extreme pH, high temperature etc). Some bacteria may switch to a resting state by forming spores.
Endospores
Endospores are formed, as the name suggests, inside the bacterial cell. Endospores are highly resistant and can be formed by members of the phylum Bacillota. The genera, which are most important in veterinary medicine are Bacillus and the closely related genus Paenibacillus as well as Clostridium and the closely related genera Paeniclostridium and Clostridioides. In each bacterial cell only one spore is formed and it can survive without access to nutrients. Endospores can withstand high and low temperatures, dehydration, chemical disinfectants and UV radiation, and the reason why they are so resistant is that they contain almost no water, and that the cell wall contains dipicolinic acid. The spores also contains much calcium (Ca) and when Ca2+ is pumped into the spore, the water is pumped out. In addition to the envelope (cortex) the spore contains primarily just DNA, ribosomes and polymerases. This is sufficient for the spores to germinate i.e. develop into bacteria again when the conditions become favorable.
Treatment with moist heat (+121°C) for 15 minutes is required to kill endospores, since they are so resistant.
Use in diagnostics
Endospores may be of importance for identification of bacteria. The position of the spore in the mother cell can give information about possible bacteria. B. cereus, B. subtilis and C. tetani for instance, have a central, subterminal and terminal spore, respectively.
Länk
Video clip at YouTube that is recommended: "Bacterial Spore Formation".
Akinetes
The orders Nostocales and Stigonematales of the phylum Cyanobacteria form so-called akinetes, which is a type of resting stage for these bacteria. Akinetes is not the same as endospores and they are not as resistant, but in the form of akineter these cyanobacteria survive the cold (winter season) and dehydration.
Exospores
The genera Actinomyces and Streptomyces of the phylum Actinomycetota can form a type of spores referred to as exospores. Exospores are formed by budding of the mycelium like structures (filaments), which these bacteria exhibit during growth. Exospores are not as resistant as endospores and therefore not of the same clinical significance. They also have a completely different structure.
VBNC
Is actually something different from spores, see VBNC (Viable but nonculturable) below.
Updated: 2023-03-08.
Streaking
 Streaking is a technique used in bacteriology for isolating individual bacterial species or strains from a sample containing a mixed bacterial flora. A sterile inoculation loop (plastic or platinum), which has been dipped into the sample, is used to streak on 1/3 (alt. 1/4) of an agar plate. Then you take a new loop and streak from the first third to the next third of the plate. This is then repeated once (or twice). In this way you get a significant decrease of bacteria between each third (or fourth) of the plate and the chance is good that somewhere on the plate, you can find individual colonies of the bacterium or bacteria you want to isolate. The panel to the right shows a streaking of Staphylococcus aureus subsp. aureus in three steps on a bovine blood agar plate. The two images (A and B) show the same plate photographed with lighting from above and in backlight, respectively. Note that on plate B it is possible to see the traces of the streaks even when bacterial colonies cannot be observed.
Streaking is a technique used in bacteriology for isolating individual bacterial species or strains from a sample containing a mixed bacterial flora. A sterile inoculation loop (plastic or platinum), which has been dipped into the sample, is used to streak on 1/3 (alt. 1/4) of an agar plate. Then you take a new loop and streak from the first third to the next third of the plate. This is then repeated once (or twice). In this way you get a significant decrease of bacteria between each third (or fourth) of the plate and the chance is good that somewhere on the plate, you can find individual colonies of the bacterium or bacteria you want to isolate. The panel to the right shows a streaking of Staphylococcus aureus subsp. aureus in three steps on a bovine blood agar plate. The two images (A and B) show the same plate photographed with lighting from above and in backlight, respectively. Note that on plate B it is possible to see the traces of the streaks even when bacterial colonies cannot be observed.
A colony, originating from a streaking, generally form a clone because most probably it originates from a single bacterial cell. There are, however, exceptions because some bacteria grow in the form of aggregates rather than as single cells. For bacteria that tend to swarm, plates which reduces the risk (for example, CLED agar for soma bacteria) should be used.
Updated: 2013-03-05.
Swarming
Proteus vulgaris, on a blood agar plate incubated 24h at 37°C. The colonies are spreading on the surface of the agar due to P. vulgaris are svarming.
Credit: Ingrid Hansson (BVF, SLU) & Lise-Lotte Fernström (BVF, SLU)
A swarming bacterium does not form discrete colonies on an agar plate, but flows out over the entire agar surface at a rate of 2–10 μm/s. If a bacterium swarms, one cannot determine its macromorphology, nor could colonies be counted. Swarming can be prevented for some bacteria by growing them on a medium with low electrolyte content (= low salt concentration), such as e.g. CLED agar. A variant of MacConkey-agar without NaCl can also be used to avoid swarming. Bacteria that can swarm are commonly found in the genera: Aeromonas, Bacillus, Brachyspira, Campylobacter, Clostridium, Escherichia, Paeniclostridium, Proteus, Pseudomonas, Salmonella, Serratia, Vibrio and Yersinia. It is not known why some bacteria swarm, but it has been suggested that it is to move quickly to areas with less competition for nutrients. Swarming bacteria usually have more flagella per unit area than planktonic (freely floating) bacteria. It has been observed that actively swarming bacteria of Salmonella Typhimurium exhibit an increased resistance to certain antibiotics as compared to non-swarming cells.
Updated: 2020-01-27.
Taxonomy and Phylogeny of bacteria
Introduction
| Rank | Suffix | Example |
| Subspecies | - | Campylobacter jejuni subsp. jejuni |
| Species | - | Campylobacter jejuni |
| Genus | - | Campylobacter |
| Family | -aceae | Campylobacteraceae |
| Order | -ales | Campylobacterales |
| Class | -ia | Campylobacteria |
| Phylum | -ota | Campylobacterota |
| Domain | - | Bacteria |
| All living organisms | - | - |
Bacterial taxonomy means naming and classification into various hierarchical groups with increased containment. I.e. a genus may consist of several different species, a family may consist of several different genera, etc. You can usually recognize the rank by its ending, i.e. if the word ends in e.g. -ales, one knows that it is the order that is intended. However, this has not been consistently implemented for all bacterial species. By the phylogeny of bacteria is meant their natural relationship, i.e. their evolutionary history
Taxonomic division
All living organisms are usually divided into three domains: Eukaryotes (Eukarya), bacteria (Bacteria) and archaea (Archaea). There are also other ways of dividing organisms, such as e.g. in seven different kingdoms: Bacteria (Bacteria), Archaea (Archaea), Protozoa (Protozoa), Chromists (Chromista), Plants (Plantae), Fungi (Fungi) and Animals (Animalia). Some bacterial groups are also divided into subfamilies, suborders and/or subclasses. Today, most scientists (at least bacteriologists) have accepted the division of organisms into three different domains and it was Carl Woese (the father of molecular phylogeny), who introduced this system in 1990 (see ref. 1 below). Now, efforts are being made for the taxonomy of the bacteria to reflect their evolutionary history (phylogeny) and as more knowledge is gained about bacteria and their phylogeny, their taxonomy will be revised.
Fig. 139:1. Carl Woese's well-known phylogenetic tree - "Tree of Life". CC BY-SA 3.0 (see ref. 1 below). This tree is based on sequence data from 16S rRNA. - Click on the image to enlarge it.
Phylogeny
The phylogeny of bacteria means their natural relationships, i.e. their evolutionary history. When bacteria began to be divided into different taxonomic categories, the classification was based on a few characteristics (e.g. gram stain, biochemical tests, sporulation ability, pathogenicity, etc.). The more features that were used, the better the division reflected the phylogeny of the bacteria. However, these features provided only blunt tools for studying the phylogeny of bacteria. The real breakthrough came when DNA sequence data was used to study the relatedness of bacteria. It was long standard to base phylogenetic trees on sequence data from 16S rRNA genes. Today, however, it has become so cheap to do whole-genome sequencing of bacteria, so you can instead base your phylogenetic trees on sequence data from large parts of the genome. Then you have access to up to 10,000 times more data compared to sequence data from the 16S rRNA gene alone, since each nucleotide position can be considered a unique and well-defined character.
Phylogenetic trees
Phylogenetic trees (or family trees), based on sequence data, are calculated by using special computer programs, which can handle large amounts of data. In the type of phylogenetic trees shown on VetBact, the horizontal distances are proportional to the sequence differences in the compared genes. Thus the distances between two organisms are shorter, the closer they are related to each other.
Phylum (pl. phyla)
The rules for naming bacteria [International Code of Nomenclature of Prokaryotes (ICNP)] did not previously include the taxonomic category (rank) phylum, but recently (February 2021) the members of the International Committee on Systematics of Prokaryotes (ICSP) have decided that from now on it will be so. It has been decided that all names of phyla should be written in italics (which has been done at VetBact also before) and have the suffix -ota. In addition, the phylum name must be based on a genus, which should constitute the nomenclature type (see ref. 2 below).
It is usually said that the phyla of bacteria form their main lineages of evolution and now there are over 40 described phyla. Eleven of these phyla are represented in VetBact. The largest phylum is Pseudomonadota (formerly Proteobacteria) and it is also the phylum that contains the greatest number of pathogenic bacteria. In VetBact there are currently 92 bacterial species, which belong to the phylum Pseudomonadota.
References
1. Woese CR, Kandler O & Wheelis ML. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. PNAS 87(12):4576–4579.
2. Oren A, & Garrity GM (2021). Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 71(10):005056.
Updated: 2023-04-17.
Taxonomy of mollicutes (mycoplasmas)
Fig. 136:1. The new taxonomy of the mollicutes (Mycoplasmas). Red taxa represent new names. Green names represent taxa where the names have not been changed and which belong to the same higher taxon as before. Yellow names represent taxa where the names have not been changed, but which have been placed within another higher taxon than before. Genera of importance within veterinary medicine are underlined. Note that not all higher taxa within the phylum Mycoplasmatota are included in this figure. - Click on the image to enlarge it.
Fig. 136:2. Species of mollicutes (mycoplasmas) with importance for veterinary medicine. At the top of each box is the name of the different genera and below the genus name follow the different species names. The same color coding is used in Fig. 2 as in Fig. 1. All species in the figure are included in VetBact, but A. laidlawii does not have much veterinary significance. - Click on the image to enlarge it.
Introduction
The taxonomy of mollicutes (mycoplasmas) has previously been controversial because it has not reflected their phylogeny, i.e. their natural relations. A Canadian and an Israeli research group have now carried out a comprehensive study based on whole-genome sequencing, where a thorough revision of the taxonomy of the mollicutes has also been proposed (see reference 1 below). Later, new phylum names were also introduced and the mollicutes now belong to the phylum Mycoplasmatota, which was previously called Tenericutes (see reference 2 below).
The revised taxonomy
In the revised taxonomy, a new order has been introduced, namely Mycoplasmoidales, which includes the new families Mycoplasmoidaceae and Metamycoplasmataceae. The family Mycoplasmoidaceae contains the new genera Malacoplasma and Mycoplasmoides as well as the genera Eperythrozoon and Ureaplasma. The genus Eperythrozoon was previously tentatively transferred to the genus Mycoplasma when it became clear that these organisms are not rickettsiae. However, the genus name Eperythrozoon has priority and therefore these organisms should still have the original genus name, even though they belong to the class Mollicutes. The family Metamycoplasmataceae contains the new genera Mesomycoplasma, Metamycoplasma and Mycoplasmopsis.
The only previously called mycoplasmas, which still belong to the order Mycoplasmatales, are those that belong to the so-called the mycoides group. This is good, because in that group one finds the most important "mycoplasmas" from a veterinary and global point of view. Now you should only use the term "mycoplasma" as a trivial name if you are referring to organisms, which belong to the revised genus Mycoplasma. Otherwise, the term "mollicutes" should be used. The genera Spiroplasma, Entomoplasma and Mesoplasma formerly belonged to the order Entomoplasmatales. However, to emphasize the close relationship with the mycoplasmas of the mycoides group, they should be placed in the order Mycoplasmatales. The genera Spiroplasma, Entomoplasma and Mesoplasma have hardly any significance in veterinary medicine, but the latter two should perhaps be placed in the two families (Entomoplasmataceae and Mesoplasmataceae, respectively) under the order Mycoplasmatales.
Summary
The new taxonomy for the mollicutes will probably create some misunderstandings and cause confusion because the five new family names are so similar to each other. In the long run, however, the new taxonomy will be a support for those who are interested in the connections between the various organisms within this interesting bacterial group.
References
1. Gupta RS, Sawnani S, Adeolu M et al. 2018. Phylogenetic framework for the phylum Tenericutes based on genome sequence data: proposal for the creation of a new order Mycoplasmoidales ord. nov., containing two new families Mycoplasmoidaceae fam. nov. and Metamycoplasmataceae fam. nov. harbouring Eperythrozoon, Ureaplasma and five novel genera. Antonie van Leeuwenhoek 111:1583–1630.
2. Oren A & Garrity GM. 2021. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 71(10):005056.
Updated: 2023-04-12.
Toxins
Introduction
Bacterial toxins are important virulence factors and they are usually classified as exotoxins (exo = outside) or endotoxins (endo = inside). Exotoxins are actively synthesized and secreted, whereas endotoxins are part of the bacteria and are released upon bacterial death and lysis. Most endotoxins are localized to the bacterial envelope and often, only lipopolysaccharides (LPS) of the outer membrane of gram negative bacteria are affiliated to thid group (see also lipopolysaccharide above). Endotoxins are heat stable.
Exotoxins
Exotoxins are typically heat sensitive (heat labile) proteins, but some are heat stable polypeptides. Exotoxins may be formed by both gram positive and gram negative bacteria. Among bacterial exotoxins, some of natures most potent toxins are found. The lethal dose for humans of e.g. the botulinum toxin is only 1-2 ng (1 ng = 0,000 000 001 g) per kg body weight at intraveneous administration. Thus, the botulinum toxin is approximately 6 000 000 times more toxic than the rattle snake venom.
Exotoxins can in turn be classified in various ways, e.g. according to:
- The tissue they affect: e.g. enterotoxins, hepatotoxins, leukocidins and neurotoxins.
- The bacterium that produces the toxin: e.g. anthrax toxin, botulinum toxin, cyanotoxin, shiga toxin, staphylococcal toxin and tetanus toxin.
- The structure of the toxin: e.g. AB toxins, which consist of two different protein subunits, A and B. A and B refers to active and binding, respectively, i.e. the actions of the subunits on the target cell. There are also AB5-toxins, which consist of 5 identical subunits (B) and one unique subunit (A), which has a different structure.
- The action of the toxin: e.g. invasins, hemolysins, toxins with enzymatic activity and channel forming toxins. Invasins make it easier for the bacteria to invade the tissue of the host animal and hemolysins can lyse the erythrocytes (and other cells) of the host animal (see hemolysis above). Toxins with enzymatic activity can be lipases, which split phospholipids in the cell membrane which in turn will cause the host cell to be fragmented (see also hemolysis above). Channel forming toxins (= porins) will get the ion gradient over the cell membrane to collapse and that will also cause fragmentation of the host cell (see also hemolysis above).
- The target organelle within or outside the host cell may be e.g. extracellular matrix, the plasma membrane, the cytoplasm and the immune system. Toxins targeting extracellular matrix are often enzymes (hyaluronidase, collagenase and elastase etc.), which can digest the constituents. Toxins targeting the plasma membrane affect its permeability and interfere with its transmembrane signaling system. Toxins targeting components in the cytoplasm also interfere with signaling systems or cytoskeleton. Toxins that cause dysfunctions in the immune system are so-called superantigens.
The different classification systems are unfortunately a source of confusion, because one toxin can have different names depending on the applied classification system. The botulinum toxin e.g. is both a neurotoxin and an AB toxin. Abbreviations are often used and the botulinum toxin for instance can be designated BoNT.
A toxoid is a protein toxin, which has been denatured (by e.g. heat or chemical treatment) and has, therefore, lost its toxicity. A toxoid retains, however, its antigenic properties and can, therefore, be used in vaccines.
Endotoxins
LPS does not exist in gram positive bacteria, but except LPS, there are some other endotoxins and one example is the Cry protein in Bacillus thuringiensis, which is a so-called δ-endotoxin. Endotoxins are in general much less toxic than exotoxins.
Secretion systems
Bacteria have complicated secretion systems to be able to export proteins to the surrounding or to the cytoplasm of cells of the host animal. Proteins, which are exported are often toxins. Information about secretion systems have also been included in the term list. See Secretion system, bacterial.
Updated: 2019-01-25.
VBNC (Viable but nonculturable)
VBNC refers to bacteria that are viable but not culturable. These bacteria have due to adverse conditions entered into a state of very low metabolic activity, and therefore they do not divide any longer. This condition is not as resistant as spores, but they can exist as VBNC for at least one year. Under the right conditions, VBNC bacteria may return to the normal state and become culturable again.Examples of bacterial genera and species, which may enter into the VBNC state: Aeromonas, Burkholderia, Campylobacter, Enterobacter, Entercoccus, Escherichia coli, Francisella, Helicobacter, Klebsiella, Legionella, Listeria, Mycobacterium, Pasteurella, Pseudomonas, Salmonella, Serratia, Shigella, Streptococcus and Vibrio.
Updated: 2013-02-26.
Virulence and virulence factors
Introduction
Virulence is not synonymous with pathogenicity (see also pathogenicity above), but describes the degree of damage that the pathogen has caused the host animal. A highly virulent bacterium is very contagious and/or gives severe symptoms. Thus, the infectious dose (ID) is lower for a high-virulent than for a low-virulent microorganism.
Virulence factors
Virulence factors are the components that bacteria produce in order to:
- Colonize some tissue of the host animal (adhesins).
- Invade tissues (tissue-degrading enzymes, see Toxins).
- Invade and get out the cells of the host (applies for intracellularbacteria).
- Escape the immune system of the host animal (by producing a capsule or by means of phase variation, which refers to a change in expression of surface proteins).
- Inhibit the immune system of the host animal (by digesting the antibodies of the host animal with specific proteases). Some mollicutes (mycoplasmas) have a special system, called MIB-MIP (Mycoplasma Immunoglobulin Binding - Mycoplasma Immunoglobulin Protease). MIB-MIP are surface proteins that bind to antibodies and cut off the VH domain from the immunoglobulin.
- Utilize nutrients from the host cells (by means of e.g. siderophores).
- For some of the bacterial toxins (e.g. botulinum toxin and tetanus toxin), the functions for the bacteria are not known, i.e. which benefits the bacteria may have from them. However, these toxins are still very important virulence factors.
Virulence genes are the genes encoding for virulence factors or components required for the synthesis of virulence factors.
Updated: 2025-01-08.
X and V factor test
Avibacterium paragallinarum, cultivated on bovine blood agar together with a streak of Staphylococcus aureus. A. paragallinarum has increased growth with bigger colonies close to the streak of Staphylococcus aureus, which cause hemolysis and release of the V factor from the erythrocytes.
Image: Ingrid Hansson & Karl-Erik Johansson (BVF, SLU). - Click on the image to enlarge it.
Introduction
Some bacteria produce substances that other bacteria need for growth. Examples of such substances are the X factor and the V factor. The X-Factor, which is heat stable, can be protoporphyrin IX, hemin or other iron-containing porphyrins, and are found in red blood cells as well as extracellularly in blood. The X-factor, also known as Stuart-Prower Factor, plays a role in both coagulation and bacteriology. In coagulation, it is an essential clotting factor, while in bacteriology, it functions as a growth factor required by certain bacteria. The V-factor, which is a heat sensitive, may be nicotinamide adenine dinukleotide (NAD) eller nicotinamide adenine dinukleotide phosphate (NADP), and is predominantly intracellular in red blood cells. X and V factor dependence can be detected either by the X and V factor test or by a disc test and the result can provide valuable information when identifying bacteria.
X and V factor test
The X and V factor test is used to determine how a certain bacterium grows on blood agar and on a minimal medium (for instance TSA) near a streak of a bacterium that produces the V factor. First, the two agar plates are inoculated with the bacterium to be examined, and then a streak of Staphylococcus aureus is done on both plates. S. aureus produces the V-factor during growth. The plates are then incubated in 5-10% CO2 at 37°C for 2-3 days. Bacteria, which only require V factor, will form larger colonies (called satellite colonies) around the S. aureus streak on both plates, while bacteria that require both V and X factors will only grow on the blood agar plate and give larger satellite colonies around the S. aureus streak.
Disc test
With a disc test, one can also investigate whether a bacterium requires the V factor or both the X and V factors. This test is carried out with three filter paper discs impregnated with V factor, X factor and a mixture of X and V factors, respectively. The discs should then be applied onto an agar plate, where the bacterium to be examined has been inoculated. This agar plate (e.g. TSA) must not contain X or V factor. The bacteria will then grow out as satellite colonies near the respective disc with the growth factor(s) that the bacterium in question needs.
Hematin agar
Hematin agar is used to cultivate fastidious bacteria and it contains blood that has been hemolysed by heating. The X-factor, which is heat stable, has thus been released from the red blood cells. The V-factor, which is a heat sensitive, has been partially destroyed, although the NAD+ nucleosidase (NADase), which is present in blood, has been inactivated by the heat.
Why do bacteria need X and V factors?
The X factor (hemin) is needed for synthesis of cytochrome C and other iron-containing respiratory enzymes. The V factor (NAD and NADP) is required for reduction-oxidation (redox) processes in the cell metabolism.
Updated: 2024-11-04.

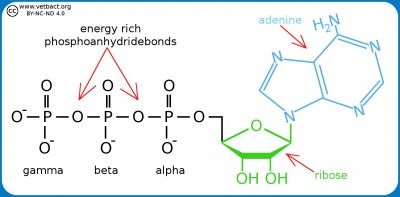
![Click to enlarge (opens in a new window). Adherence [under revision]](/images/terms/230517_Eng_v2.jpg)