Matrix-Assisted Laser Desorption/Ionization Time Of Flight Mass Spectrometry (MALDI-TOF MS)
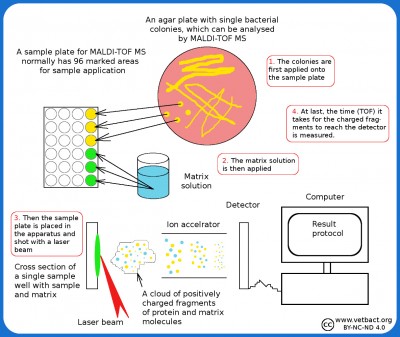
Schematic description of the principle of MALDI-TOF MS. - Click on the image to enlarge it.
The principle of MALDI-TOFMass spectrometry based on the MALDI-TOF (or Maldi only) means that the bacterial isolate to be analyzed, is adsorbed to some type of carrier material (matrix). The isolate is then irradiated with laser UV light, so that the molecules in the bacteria are broken into positively charged fragments (ionization), which are thrown towards a detector. The time it takes for the fragment to reach the detector (time of flight) is measured. The time is dependent on fragment size and charge. Also very large molecules (proteins and nucleic acids) can be fragmented and ionized in this way. Large molecules give rise to many fragments and a characteristic mass spectrum, which can be used for identification. The use of MALDI-TOF MS for identification of bacteriaOne can perform these analyzes directly on bacterial colonies and the resulting mass spectrum is then compared with stored mass spectra of known bacterial species. Thus, you will get an analytical response within a minute and the method is considered to be very reliable. The more mass spectra of known bacteria you have to compare with, the safer the method will be. MALDI-TOF MS is already used in some laboratories for veterinary bacteriology and many researchers believe that this technique will be tomorrow's routine method for identification of bacteria. The instrument is still very expensive, but material costs are low. Score valueThe software for the Maldi system contains a database with information about which fragments one can possibly get from a particular bacterium. This information can be used by the system to compare with unknown isolates and calculate a score value for each isolate. The score value is a measure of the probability that a particular isolate will represent a particular bacterial species/strain in the reference library. A score value between 0.000 and 1.699 means that identification is not possible since the peak of the unknown isolate does not represent any bacterium in the reference library. A score value between 1.700 and 1.999 are reliable only to the genus level, but a low confidence of identification at species level compared to reference peak list. . A score value between 2.000 and 3.000 are considered accurate of both genus- and species-level. Updated: 2024-12-09. |